Integrative Proteomics and Transcriptomics Profiles of the Oviduct Reveal the Prolificacy-Related Candidate Biomarkers of Goats (Capra hircus) in Estrous Periods
- PMID: 36499219
- PMCID: PMC9737051
- DOI: 10.3390/ijms232314888
Integrative Proteomics and Transcriptomics Profiles of the Oviduct Reveal the Prolificacy-Related Candidate Biomarkers of Goats (Capra hircus) in Estrous Periods
Abstract
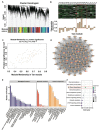
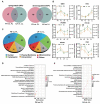
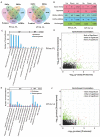
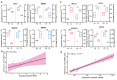
The oviduct is a dynamic reproductive organ for mammalian reproduction and is required for gamete storage, maturation, fertilization, and early embryonic development, and it directly affects fecundity. However, the molecular regulation of prolificacy occurring in estrous periods remain poorly understood. This study aims to gain a better understanding of the genes involved in regulating goat fecundity in the proteome and transcriptome levels of the oviducts. Twenty female Yunshang black goats (between 2 and 3 years old, weight 52.22 ± 0.43 kg) were divided into high- and low-fecundity groups in the follicular (FH and FL, five individuals per group) and luteal (LH and LL, five individuals per group) phases, respectively. The DIA-based high-resolution mass spectrometry (MS) method was used to quantify proteins in twenty oviducts. A total of 5409 proteins were quantified, and Weighted gene co-expression network analysis (WGCNA) determined that the tan module was highly associated with the high-fecundity trait in the luteal phase, and identified NUP107, ANXA11, COX2, AKP13, and ITF140 as hub proteins. Subsequently, 98 and 167 differentially abundant proteins (DAPs) were identified in the FH vs. FL and LH vs. LL comparison groups, respectively. Parallel reaction monitoring (PRM) was used to validate the results of the proteomics data, and the hub proteins were analyzed with Western blot (WB). In addition, biological adhesion and transporter activity processes were associated with oviductal function, and several proteins that play roles in oviductal communication with gametes or embryos were identified, including CAMSAP3, ITGAM, SYVN1, EMG1, ND5, RING1, CBS, PES1, ELP3, SEC24C, SPP1, and HSPA8. Correlation analysis of proteomics and transcriptomic revealed that the DAPs and differentially expressed genes (DEGs) are commonly involved in the metabolic processes at the follicular phase; they may prepare the oviductal microenvironment for gamete reception; and the MAP kinase activity, estrogen receptor binding, and angiotensin receptor binding terms were enriched in the luteal phase, which may be actively involved in reproductive processes. By generating the proteome data of the oviduct at two critical phases and integrating transcriptome analysis, we uncovered novel aspects of oviductal gene regulation of fecundity and provided a reference for other mammals.
Keywords: DIA proteome; WGCNA; estrous period; fecundity; goat; oviduct; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Shah A.M., Cai Y.M., Zou H.W., Zhang X.F., Wang L.Z., Xue B., Yu P.Q., Wang Z.S., Peng Q.H. Effects of supplementation of branches and leaves trimmed from tea plant on growth performance, rumen fermentation and meat composition of Nanjiang yellow goats. Animals. 2019;9:590. doi: 10.3390/ani9090590. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

