Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress
- PMID: 36499255
- PMCID: PMC9740226
- DOI: 10.3390/ijms232314926
Integrative Omics Analysis of Three Oil Palm Varieties Reveals (Tanzania × Ekona) TE as a Cold-Resistant Variety in Response to Low-Temperature Stress
Abstract
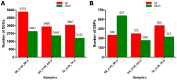
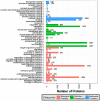
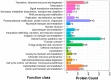
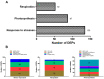
Oil palm (Elaeis guineensis Jacq.) is an economically important tropical oil crop widely cultivated in tropical zones worldwide. Being a tropical crop, low-temperature stress adversely affects the oil palm. However, integrative leaf transcriptomic and proteomic analyses have not yet been conducted on an oil palm crop under cold stress. In this study, integrative omics transcriptomic and iTRAQ-based proteomic approaches were employed for three oil palm varieties, i.e., B × E (Bamenda × Ekona), O × G (E. oleifera × Elaeis guineensis), and T × E (Tanzania × Ekona), in response to low-temperature stress. In response to low-temperature stress at (8 °C) for 5 days, a total of 5175 up- and 2941 downregulated DEGs in BE-0_VS_BE-5, and a total of 3468 up- and 2443 downregulated DEGs for OG-0_VS_OG-5, and 3667 up- and 2151 downregulated DEGs for TE-0_VS_TE-5 were identified. iTRAQ-based proteomic analysis showed 349 up- and 657 downregulated DEPs for BE-0_VS_BE-5, 372 up- and 264 downregulated DEPs for OG-0_VS_OG-5, and 500 up- and 321 downregulated DEPs for TE-0_VS_TE-5 compared to control samples treated at 28 °C and 8 °C, respectively. The KEGG pathway correlation of oil palm has shown that the metabolic synthesis and biosynthesis of secondary metabolites pathways were significantly enriched in the transcriptome and proteome of the oil palm varieties. The correlation expression pattern revealed that TE-0_VS_TE-5 is highly expressed and BE-0_VS_BE-5 is suppressed in both the transcriptome and proteome in response to low temperature. Furthermore, numerous transcription factors (TFs) were found that may regulate cold acclimation in three oil palm varieties at low temperatures. Moreover, this study identified proteins involved in stresses (abiotic, biotic, oxidative, and heat shock), photosynthesis, and respiration in iTRAQ-based proteomic analysis of three oil palm varieties. The increased abundance of stress-responsive proteins and decreased abundance of photosynthesis-related proteins suggest that the TE variety may become cold-resistant in response to low-temperature stress. This study may provide a basis for understanding the molecular mechanism for the adaptation of oil palm varieties in response to low-temperature stress in China.
Keywords: TE; cold tolerance; low-temperature stress; oil palm; proteome; transcriptome.
Conflict of interest statement
The authors declared no conflict of interest.
Figures






Similar articles
-
Multi-Omics Approaches in Oil Palm Research: A Comprehensive Review of Metabolomics, Proteomics, and Transcriptomics Based on Low-Temperature Stress.Int J Mol Sci. 2024 Jul 13;25(14):7695. doi: 10.3390/ijms25147695. Int J Mol Sci. 2024. PMID: 39062936 Free PMC article. Review.
-
Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research.Int J Mol Sci. 2024 Aug 7;25(16):8625. doi: 10.3390/ijms25168625. Int J Mol Sci. 2024. PMID: 39201312 Free PMC article. Review.
-
iTRAQ-based comparative proteomic analysis of two coconut varieties reveals aromatic coconut cold-sensitive in response to low temperature.J Proteomics. 2020 May 30;220:103766. doi: 10.1016/j.jprot.2020.103766. Epub 2020 Mar 31. J Proteomics. 2020. PMID: 32240811
-
Exploiting transcriptome data for the development and characterization of gene-based SSR markers related to cold tolerance in oil palm (Elaeis guineensis).BMC Plant Biol. 2014 Dec 19;14:384. doi: 10.1186/s12870-014-0384-2. BMC Plant Biol. 2014. PMID: 25522814 Free PMC article.
-
Correlation analysis of cold-related gene expression with physiological and biochemical indicators under cold stress in oil palm.PLoS One. 2019 Nov 27;14(11):e0225768. doi: 10.1371/journal.pone.0225768. eCollection 2019. PLoS One. 2019. PMID: 31774880 Free PMC article.
Cited by
-
Core transcriptome network modulates temperature (heat and cold) and osmotic (drought, salinity, and waterlogging) stress responses in oil palm.Front Plant Sci. 2024 Dec 23;15:1497017. doi: 10.3389/fpls.2024.1497017. eCollection 2024. Front Plant Sci. 2024. PMID: 39764238 Free PMC article.
-
Proteomics: An Essential Tool to Study Plant-Specialized Metabolism.Biomolecules. 2024 Nov 30;14(12):1539. doi: 10.3390/biom14121539. Biomolecules. 2024. PMID: 39766246 Free PMC article. Review.
-
Integrated Transcriptomic and Metabolomics Analyses Reveal Molecular Responses to Cold Stress in Coconut (Cocos nucifera L.) Seedlings.Int J Mol Sci. 2023 Sep 26;24(19):14563. doi: 10.3390/ijms241914563. Int J Mol Sci. 2023. PMID: 37834015 Free PMC article.
-
Multi-Omics Approaches in Oil Palm Research: A Comprehensive Review of Metabolomics, Proteomics, and Transcriptomics Based on Low-Temperature Stress.Int J Mol Sci. 2024 Jul 13;25(14):7695. doi: 10.3390/ijms25147695. Int J Mol Sci. 2024. PMID: 39062936 Free PMC article. Review.
-
Unveiling the Secrets of Oil Palm Genetics: A Look into Omics Research.Int J Mol Sci. 2024 Aug 7;25(16):8625. doi: 10.3390/ijms25168625. Int J Mol Sci. 2024. PMID: 39201312 Free PMC article. Review.
References
-
- Bittencourt C.B., Carvalho da Silva T.L., Rodrigues Neto J.C., Vieira L.R., Leão A.P., de Aquino Ribeiro J.A., Abdelnur P.V., de Sousa C.A.F., Souza M.T., Jr. Insights from a Multi-Omics Integration (MOI) Study in Oil Palm (Elaeis guineensis Jacq.) Response to Abiotic Stresses: Part One—Salinity. Plants. 2022;11:1755. doi: 10.3390/plants11131755. - DOI - PMC - PubMed
-
- Lei X., Xiao Y., Xia W., Mason A.S., Yang Y., Ma Z., Peng M. RNA-seq analysis of oil palm under cold stress reveals a different C-repeat binding factor (CBF) mediated gene expression pattern in Elaeis guineensis compared to other species. PLoS ONE. 2014;9:e114482. doi: 10.1371/journal.pone.0114482. - DOI - PMC - PubMed
-
- Corley R.H.V., Tinker P.B. The Oil Palm. 4th ed. Blackwell Science; Oxford, UK: 2003. Vegetative propagation and biotechnology; pp. 201–215.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

