Metabolic and Transcriptomic Profiling Reveals Etiolated Mechanism in Huangyu Tea (Camellia sinensis) Leaves
- PMID: 36499369
- PMCID: PMC9740216
- DOI: 10.3390/ijms232315044
Metabolic and Transcriptomic Profiling Reveals Etiolated Mechanism in Huangyu Tea (Camellia sinensis) Leaves
Abstract
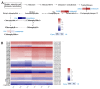
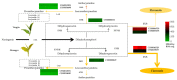
Leaf color is one of the key factors involved in determining the processing suitability of tea. It relates to differential accumulation of flavor compounds due to the different metabolic mechanisms. In recent years, photosensitive etiolation or albefaction is an interesting direction in tea research field. However, the molecular mechanism of color formation remains unclear since albino or etiolated mutants have different genetic backgrounds. In this study, wide-target metabolomic and transcriptomic analyses were used to reveal the biological mechanism of leaf etiolation for 'Huangyu', a bud mutant of 'Yinghong 9'. The results indicated that the reduction in the content of chlorophyll and the ratio of chlorophyll to carotenoids might be the biochemical reasons for the etiolation of 'Huangyu' tea leaves, while the content of zeaxanthin was significantly higher. The differentially expressed genes (DEGs) involved in chlorophyll and chloroplast biogenesis were the biomolecular reasons for the formation of green or yellow color in tea leaves. In addition, our results also revealed that the changes of DEGs involved in light-induced proteins and circadian rhythm promoted the adaptation of etiolated tea leaves to light stress. Variant colors of tea leaves indicated different directions in metabolic flux and accumulation of flavor compounds.
Keywords: Camellia sinensis cv. Huangyu; biological mechanism; chlorophyll; leaf color; yellowing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Transcriptome and Biochemical Analyses of a Chlorophyll-Deficient Bud Mutant of Tea Plant (Camellia sinensis).Int J Mol Sci. 2023 Oct 11;24(20):15070. doi: 10.3390/ijms242015070. Int J Mol Sci. 2023. PMID: 37894753 Free PMC article.
-
A Comparative Metabolomic Analysis Reveals Difference Manufacture Suitability in "Yinghong 9" and "Huangyu" Teas (Camellia sinensis).Front Plant Sci. 2021 Dec 14;12:767724. doi: 10.3389/fpls.2021.767724. eCollection 2021. Front Plant Sci. 2021. PMID: 34970283 Free PMC article.
-
Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in 'Anji Baicha' (Camellia sinensis).BMC Plant Biol. 2016 Sep 8;16(1):195. doi: 10.1186/s12870-016-0885-2. BMC Plant Biol. 2016. PMID: 27609021 Free PMC article.
-
Review: the effect of light on the key pigment compounds of photosensitive etiolated tea plant.Bot Stud. 2021 Dec 11;62(1):21. doi: 10.1186/s40529-021-00329-2. Bot Stud. 2021. PMID: 34897570 Free PMC article. Review.
-
Anthocyanin metabolism and its differential regulation in purple tea (Camellia sinensis).Plant Physiol Biochem. 2023 Aug;201:107875. doi: 10.1016/j.plaphy.2023.107875. Epub 2023 Jul 9. Plant Physiol Biochem. 2023. PMID: 37451003 Review.
Cited by
-
Light Intensity Modulates the Functional Composition of Leaf Metabolite Groups and Phyllosphere Prokaryotic Community in Garden Lettuce (Lactuca sativa L.) Plants at the Vegetative Stage.Int J Mol Sci. 2024 Jan 25;25(3):1451. doi: 10.3390/ijms25031451. Int J Mol Sci. 2024. PMID: 38338730 Free PMC article.
-
Transcriptome and Biochemical Analyses of a Chlorophyll-Deficient Bud Mutant of Tea Plant (Camellia sinensis).Int J Mol Sci. 2023 Oct 11;24(20):15070. doi: 10.3390/ijms242015070. Int J Mol Sci. 2023. PMID: 37894753 Free PMC article.
-
Metabolome profiling and transcriptome analysis unveiling the crucial role of magnesium transport system for magnesium homeostasis in tea plants.Hortic Res. 2024 Jun 3;11(7):uhae152. doi: 10.1093/hr/uhae152. eCollection 2024 Jul. Hortic Res. 2024. PMID: 38994447 Free PMC article.
-
The Chemical Composition and Transcriptome Analysis Reveal the Mechanism of Color Formation in Tea (Camellia sinensis) Pericarp.Int J Mol Sci. 2023 Aug 25;24(17):13198. doi: 10.3390/ijms241713198. Int J Mol Sci. 2023. PMID: 37686006 Free PMC article.
-
Multi-Omics Research Accelerates the Clarification of the Formation Mechanism and the Influence of Leaf Color Variation in Tea (Camellia sinensis) Plants.Plants (Basel). 2024 Jan 31;13(3):426. doi: 10.3390/plants13030426. Plants (Basel). 2024. PMID: 38337959 Free PMC article. Review.
References
-
- Yamashita H., Kambe Y., Ohshio M., Kunihiro A., Tanaka Y., Suzuki T., Nakamura Y., Morita A., Ikka T. Integrated Metabolome and Transcriptome Analyses Reveal Etiolation-Induced Metabolic Changes Leading to High Amino Acid Contents in a Light-Sensitive Japanese Albino Tea Cultivar. Front. Plant Sci. 2020;11:611140. doi: 10.3389/fpls.2020.611140. - DOI - PMC - PubMed
-
- Shin Y.H., Yang R., Shi Y.L., Li X.M., Fu Q.Y., Lu J.L., Ye J.H., Wang K.R., Ma S.C., Zheng X.Q., et al. Light-sensitive Albino Tea Plants and Their Characterization. Hortscience. 2018;53:144–147. doi: 10.21273/HORTSCI12633-17. - DOI
-
- Wu H., Shi N., An X., Liu C., Fu H., Cao L., Feng Y., Sun D., Zhang L. Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways according to Transcriptome Profiling. Int. J. Mol. Sci. 2018;19:1594. doi: 10.3390/ijms19061594. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

