SMILE Downregulation during Melanogenesis Induces MITF Transcription in B16F10 Cells
- PMID: 36499416
- PMCID: PMC9738925
- DOI: 10.3390/ijms232315094
SMILE Downregulation during Melanogenesis Induces MITF Transcription in B16F10 Cells
Abstract
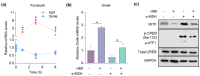
SMILE (small heterodimer partner-interacting leucine zipper protein) is a transcriptional corepressor that potently regulates various cellular processes such as metabolism and growth in numerous tissues. However, its regulatory role in skin tissue remains uncharacterized. Here, we demonstrated that SMILE expression markedly decreased in human melanoma biopsy specimens and was inversely correlated with that of microphthalmia-associated transcription factor (MITF). During melanogenesis, α-melanocyte-stimulating hormone (α-MSH) induction of MITF was mediated by a decrease in SMILE expression in B16F10 mouse melanoma cells. Mechanistically, SMILE was regulated by α-MSH/cAMP/protein kinase A signaling and suppressed MITF promoter activity via corepressing transcriptional activity of the cAMP response element-binding protein. Moreover, SMILE overexpression significantly reduced α-MSH-induced MITF and melanogenic genes, thereby inhibiting melanin production in melanocytes. Conversely, SMILE inhibition increased the transcription of melanogenic genes and melanin contents. These results indicate that SMILE is a downstream effector of cAMP-mediated signaling and is a critical factor in the regulation of melanogenic transcription; in addition, they suggest a potential role of SMILE as a corepressor in skin pigmentation.
Keywords: MITF; SMILE; cAMP; melanogenesis; skin pigmentation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Interruption of p38MAPK-MSK1-CREB-MITF-M pathway to prevent hyperpigmentation in the skin.Int J Biol Sci. 2024 Feb 17;20(5):1688-1704. doi: 10.7150/ijbs.93120. eCollection 2024. Int J Biol Sci. 2024. PMID: 38481807 Free PMC article.
-
Sargaquinoic acid ameliorates hyperpigmentation through cAMP and ERK-mediated downregulation of MITF in α-MSH-stimulated B16F10 cells.Biomed Pharmacother. 2018 Aug;104:582-589. doi: 10.1016/j.biopha.2018.05.083. Epub 2018 May 25. Biomed Pharmacother. 2018. PMID: 29803170
-
Inhibition of α-melanocyte-stimulating hormone-induced melanogenesis and molecular mechanisms by polyphenol-enriched fraction of Tagetes erecta L. flower.Phytomedicine. 2024 Apr;126:155442. doi: 10.1016/j.phymed.2024.155442. Epub 2024 Feb 15. Phytomedicine. 2024. PMID: 38394730
-
[Regulation of melanogenesis: the role of cAMP and MITF].Postepy Hig Med Dosw (Online). 2012 Jan 30;66:33-40. Postepy Hig Med Dosw (Online). 2012. PMID: 22371403 Review. Polish.
-
Involvement of adenylate cyclase/cAMP/CREB and SOX9/MITF in melanogenesis to prevent vitiligo.Mol Cell Biochem. 2021 Mar;476(3):1401-1409. doi: 10.1007/s11010-020-04000-5. Epub 2021 Jan 3. Mol Cell Biochem. 2021. PMID: 33389492 Review.
Cited by
-
Emerging Role of SMILE in Liver Metabolism.Int J Mol Sci. 2023 Feb 2;24(3):2907. doi: 10.3390/ijms24032907. Int J Mol Sci. 2023. PMID: 36769229 Free PMC article. Review.
-
Small Peptide Derived from SFRP5 Suppresses Melanogenesis by Inhibiting Wnt Activity.Curr Issues Mol Biol. 2024 May 29;46(6):5420-5435. doi: 10.3390/cimb46060324. Curr Issues Mol Biol. 2024. PMID: 38920996 Free PMC article.
References
-
- Jablonski N.G. The evolution of human skin and skin color. Annu. Rev. Anthropol. 2004;33:585–623. doi: 10.1146/annurev.anthro.33.070203.143955. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

