A Dual Role for FADD in Human Precursor T-Cell Neoplasms
- PMID: 36499482
- PMCID: PMC9738522
- DOI: 10.3390/ijms232315157
A Dual Role for FADD in Human Precursor T-Cell Neoplasms
Abstract
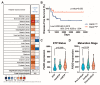
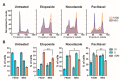
A reduction in FADD levels has been reported in precursor T-cell neoplasms and other tumor types. Such reduction would impact on the ability of tumor cells to undergo apoptosis and has been associated with poor clinical outcomes. However, FADD is also known to participate in non-apoptotic functions, but these mechanisms are not well-understood. Linking FADD expression to the severity of precursor T-cell neoplasms could indicate its use as a prognostic marker and may open new avenues for targeted therapeutic strategies. Using transcriptomic and clinical data from patients with precursor T-cell neoplasms, complemented by in vitro analysis of cellular functions and by high-throughput interactomics, our results allow us to propose a dual role for FADD in precursor T-cell neoplasms, whereby resisting cell death and chemotherapy would be a canonical consequence of FADD deficiency in these tumors, whereas deregulation of the cellular metabolism would be a relevant non-canonical function in patients expressing FADD. These results reveal that evaluation of FADD expression in precursor T-cell neoplasms may aid in the understanding of the biological processes that are affected in the tumor cells. The altered biological processes can be of different natures depending on the availability of FADD influencing its ability to exert its canonical or non-canonical functions. Accordingly, specific therapeutic interventions would be needed in each case.
Keywords: FADD; T-cell acute lymphoblastic leukemia/T-cell lymphoblastic lymphoma (T-ALL/LBL); canonical and non-canonical functions; interactomics; precursor T-cell neoplasms; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Marin-Rubio J.L., de Arriba M.C., Cobos-Fernandez M.A., Gonzalez-Sanchez L., Ors I., Sastre I., Fernandez-Piqueras J., Villa-Morales M. Deregulated FADD expression and phosphorylation in T-cell lymphoblastic lymphoma. Oncotarget. 2016;7:61485–61499. doi: 10.18632/oncotarget.11370. - DOI - PMC - PubMed
-
- Patel S., Murphy D., Haralambieva E., Abdulla Z.A., Wong K.K., Chen H., Gould E., Roncador G., Hatton C.S., Anderson A.P., et al. Increased Expression of Phosphorylated FADD in Anaplastic Large Cell and Other T-Cell Lymphomas. Biomark. Insights. 2014;9:77–84. doi: 10.4137/BMI.S16553. - DOI - PMC - PubMed
-
- Cimino Y., Costes A., Damotte D., Validire P., Mistou S., Cagnard N., Alifano M., Regnard J.F., Chiocchia G., Sautes-Fridman C., et al. FADD protein release mirrors the development and aggressiveness of human non-small cell lung cancer. Br. J. Cancer. 2012;106:1989–1996. doi: 10.1038/bjc.2012.196. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- SAF2015-70561/Ministry of Economy, Industry and Competitiveness
- B2017/BMD-3778; LINFOMAS-CM/Comunidad de Madrid
- ACCI-CIBERER-17 ISCIII/Instituto de Salud Carlos III
- FPU13/00338/Ministerio de Educación Cultura y Deporte
- JLMR/Federation of European Biochemical Societies
- JCSTF-161105/Journal of Cell Science
- EACR, Ref. #571/European Association for Cancer Research
- RTI2018-093330-B-I00/Spanish Ministry of Science, Innovation and Universities
- CIVP19S7917/Fundación Ramón Areces
- AECC, 2018; PROYE18054PIRI/Asociación Española Contra el Cáncer
- PI20/00590/Ministry of Economy, Industry and Competitiveness
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

