Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea
- PMID: 36499570
- PMCID: PMC9739956
- DOI: 10.3390/ijms232315243
Assembly and Annotation of Red Spruce (Picea rubens) Chloroplast Genome, Identification of Simple Sequence Repeats, and Phylogenetic Analysis in Picea
Abstract
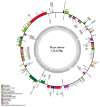
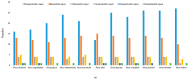
We have sequenced the chloroplast genome of red spruce (Picea rubens) for the first time using the single-end, short-reads (44 bp) Illumina sequences, assembled and functionally annotated it, and identified simple sequence repeats (SSRs). The contigs were assembled using SOAPdenovo2 following the retrieval of chloroplast genome sequences using the black spruce (Picea mariana) chloroplast genome as the reference. The assembled genome length was 122,115 bp (gaps included). Comparatively, the P. rubens chloroplast genome reported here may be considered a near-complete draft. Global genome alignment and phylogenetic analysis based on the whole chloroplast genome sequences of Picea rubens and 10 other Picea species revealed high sequence synteny and conservation among 11 Picea species and phylogenetic relationships consistent with their known classical interrelationships and published molecular phylogeny. The P. rubens chloroplast genome sequence showed the highest similarity with that of P. mariana and the lowest with that of P. sitchensis. We have annotated 107 genes including 69 protein-coding genes, 28 tRNAs, 4 rRNAs, few pseudogenes, identified 42 SSRs, and successfully designed primers for 26 SSRs. Mononucleotide A/T repeats were the most common followed by dinucleotide AT repeats. A similar pattern of microsatellite repeats occurrence was found in the chloroplast genomes of 11 Picea species.
Keywords: Pinaceae; comparative genome analysis; conifers; genome assembly and annotation; genome sequences; microsatellites; organellar genome; phylogeny; plastid genome.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Palmer J.D. Chloroplast DNA and molecular phylogeny. Bioessays. 1985;2:263–267. doi: 10.1002/bies.950020607. - DOI
-
- Mereschkowsky C. Uber natur und ursprung der chromatophoren im pflanzenreiche. Biol. Cent. 1905;25:293–604.
-
- Mereschkowsky C. Theorie der zwei Plasmaarten als Grundlage der Symbiogenesis, einer neuen Lehre von der Entstehung der Organismen. Biol. Cent. 1910;30:278–288.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

