Pharmacological Mechanism of NRICM101 for COVID-19 Treatments by Combined Network Pharmacology and Pharmacodynamics
- PMID: 36499711
- PMCID: PMC9740625
- DOI: 10.3390/ijms232315385
Pharmacological Mechanism of NRICM101 for COVID-19 Treatments by Combined Network Pharmacology and Pharmacodynamics
Abstract
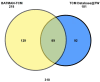
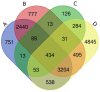
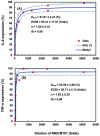
Symptom treatments for Coronavirus disease 2019 (COVID-19) infection and Long COVID are one of the most critical issues of the pandemic era. In light of the lack of standardized medications for treating COVID-19 symptoms, traditional Chinese medicine (TCM) has emerged as a potentially viable strategy based on numerous studies and clinical manifestations. Taiwan Chingguan Yihau (NRICM101), a TCM designed based on a medicinal formula with a long history of almost 500 years, has demonstrated its antiviral properties through clinical studies, yet the pharmacogenomic knowledge for this formula remains unclear. The molecular mechanism of NRICM101 was systematically analyzed by using exploratory bioinformatics and pharmacodynamics (PD) approaches. Results showed that there were 434 common interactions found between NRICM101 and COVID-19 related genes/proteins. For the network pharmacology of the NRICM101, the 434 common interacting genes/proteins had the highest associations with the interleukin (IL)-17 signaling pathway in the Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Moreover, the tumor necrosis factor (TNF) was found to have the highest association with the 30 most frequently curated NRICM101 chemicals. Disease analyses also revealed that the most relevant diseases with COVID-19 infections were pathology, followed by cancer, digestive system disease, and cardiovascular disease. The 30 most frequently curated human genes and 2 microRNAs identified in this study could also be used as molecular biomarkers or therapeutic options for COVID-19 treatments. In addition, dose-response profiles of NRICM101 doses and IL-6 or TNF-α expressions in cell cultures of murine alveolar macrophages were constructed to provide pharmacodynamic (PD) information of NRICM101. The prevalent use of NRICM101 for standardized treatments to attenuate common residual syndromes or chronic sequelae of COVID-19 were also revealed for post-pandemic future.
Keywords: NRICM101; biomarker; pharmacodynamics; pharmacogenomics; protein–protein interaction; traditional Chinese medicine.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Taiwan Chingguan Yihau may improve post-COVID-19 respiratory complications through PI3K/AKT, HIF-1, and TNF signaling pathways revealed by network pharmacology analysis.Mol Divers. 2025 Jun;29(3):2305-2321. doi: 10.1007/s11030-024-10993-8. Epub 2024 Oct 9. Mol Divers. 2025. PMID: 39382736
-
In Silico and In Vitro studies of taiwan chingguan yihau (NRICM101) on TNF-α/IL-1β-induced human lung cells.Biomedicine (Taipei). 2022 Sep 1;12(3):56-71. doi: 10.37796/2211-8039.1378. eCollection 2022. Biomedicine (Taipei). 2022. PMID: 36381194 Free PMC article.
-
A traditional Chinese medicine formula NRICM101 to target COVID-19 through multiple pathways: A bedside-to-bench study.Biomed Pharmacother. 2021 Jan;133:111037. doi: 10.1016/j.biopha.2020.111037. Epub 2020 Nov 19. Biomed Pharmacother. 2021. PMID: 33249281 Free PMC article.
-
Chinese herbal prescriptions for COVID-19 management: Special reference to Taiwan Chingguan Yihau (NRICM101).Front Pharmacol. 2022 Oct 5;13:928106. doi: 10.3389/fphar.2022.928106. eCollection 2022. Front Pharmacol. 2022. PMID: 36278162 Free PMC article. Review.
-
Potential effect of Maxing Shigan decoction against coronavirus disease 2019 (COVID-19) revealed by network pharmacology and experimental verification.J Ethnopharmacol. 2021 May 10;271:113854. doi: 10.1016/j.jep.2021.113854. Epub 2021 Jan 26. J Ethnopharmacol. 2021. PMID: 33513419 Free PMC article.
Cited by
-
NRICM101 in combatting COVID-19 induced brain fog: Neuroprotective effects and neurovascular integrity preservation in hACE2 mice.J Tradit Complement Med. 2024 Jul 3;15(1):36-50. doi: 10.1016/j.jtcme.2024.07.001. eCollection 2025 Jan. J Tradit Complement Med. 2024. PMID: 39807266 Free PMC article.
-
Bioinformatics-based investigation on the genetic influence between SARS-CoV-2 infections and idiopathic pulmonary fibrosis (IPF) diseases, and drug repurposing.Sci Rep. 2023 Mar 22;13(1):4685. doi: 10.1038/s41598-023-31276-6. Sci Rep. 2023. PMID: 36949176 Free PMC article.
-
ACE2 and a Traditional Chinese Medicine Formula NRICM101 Could Alleviate the Inflammation and Pathogenic Process of Acute Lung Injury.Medicina (Kaunas). 2023 Aug 26;59(9):1554. doi: 10.3390/medicina59091554. Medicina (Kaunas). 2023. PMID: 37763673 Free PMC article.
-
Computational identification of differentially-expressed genes as suggested novel COVID-19 biomarkers: A bioinformatics analysis of expression profiles.Comput Struct Biotechnol J. 2023;21:3339-3354. doi: 10.1016/j.csbj.2023.06.007. Epub 2023 Jun 12. Comput Struct Biotechnol J. 2023. PMID: 37347079 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

