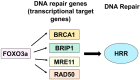
FOXO3a Mediates Homologous Recombination Repair (HRR) via Transcriptional Activation of MRE11, BRCA1, BRIP1, and RAD50
- PMID: 36500714
- PMCID: PMC9741359
- DOI: 10.3390/molecules27238623
FOXO3a Mediates Homologous Recombination Repair (HRR) via Transcriptional Activation of MRE11, BRCA1, BRIP1, and RAD50
Abstract
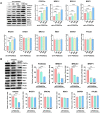
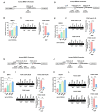
To test whether homologous recombination repair (HRR) depends on FOXO3a, a cellular aging model of human dermal fibroblast (HDF) and tet-on flag-h-FOXO3a transgenic mice were studied. HDF cells transfected with over-expression of wt-h-FOXO3a increased the protein levels of MRE11, BRCA1, BRIP1, and RAD50, while knock-down with siFOXO3a decreased them. The protein levels of MRE11, BRCA1, BRIP1, RAD50, and RAD51 decreased during cellular aging. Chromatin immunoprecipitation (ChIP) assay was performed on FOXO3a binding accessibility to FOXO consensus sites in human MRE11, BRCA1, BRIP1, and RAD50 promoters; the results showed FOXO3a binding decreased during cellular aging. When the tet-on flag-h-FOXO3a mice were administered doxycycline orally, the protein and mRNA levels of flag-h-FOXO3a, MRE11, BRCA1, BRIP1, and RAD50 increased in a doxycycline-dose-dependent manner. In vitro HRR assays were performed by transfection with an HR vector and I-SceI vector. The mRNA levels of the recombined GFP increased after doxycycline treatment in MEF but not in wt-MEF, and increased in young HDF comparing to old HDF, indicating that FOXO3a activates HRR. Overall, these results demonstrate that MRE11, BRCA1, BRIP1, and RAD50 are transcriptional target genes for FOXO3a, and HRR activity is increased via transcriptional activation of MRE11, BRCA1, BRIP1, and RAD50 by FOXO3a.
Keywords: BRCA1; BRIP1; FOXO3a; MRE11; RAD50; homologous recombination repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


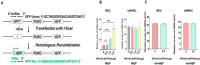
Similar articles
-
The Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex promotes trinucleotide repeat expansions independently of homologous recombination.DNA Repair (Amst). 2016 Jul;43:1-8. doi: 10.1016/j.dnarep.2016.04.012. Epub 2016 May 2. DNA Repair (Amst). 2016. PMID: 27173583
-
Triapine disrupts CtIP-mediated homologous recombination repair and sensitizes ovarian cancer cells to PARP and topoisomerase inhibitors.Mol Cancer Res. 2014 Mar;12(3):381-393. doi: 10.1158/1541-7786.MCR-13-0480. Epub 2014 Jan 10. Mol Cancer Res. 2014. PMID: 24413181 Free PMC article.
-
Aberrations of the MRE11-RAD50-NBS1 DNA damage sensor complex in human breast cancer: MRE11 as a candidate familial cancer-predisposing gene.Mol Oncol. 2008 Dec;2(4):296-316. doi: 10.1016/j.molonc.2008.09.007. Epub 2008 Oct 7. Mol Oncol. 2008. PMID: 19383352 Free PMC article.
-
The MRE11 complex: A versatile toolkit for the repair of broken DNA.DNA Repair (Amst). 2020 Jul-Aug;91-92:102869. doi: 10.1016/j.dnarep.2020.102869. Epub 2020 May 15. DNA Repair (Amst). 2020. PMID: 32480356 Review.
-
Human syndromes with genomic instability and multiprotein machines that repair DNA double-strand breaks.Histol Histopathol. 2003 Jan;18(1):225-43. doi: 10.14670/HH-18.225. Histol Histopathol. 2003. PMID: 12507302 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

