Phylogenetic and Phylodynamic Analyses of Soybean Mosaic Virus Using 305 Coat Protein Gene Sequences
- PMID: 36501296
- PMCID: PMC9736121
- DOI: 10.3390/plants11233256
Phylogenetic and Phylodynamic Analyses of Soybean Mosaic Virus Using 305 Coat Protein Gene Sequences
Abstract
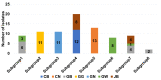
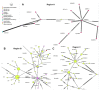
Soybean mosaic virus (SMV) of the family Potyviridae is the most devastating virus that infects soybean plants. In this study, we obtained 83 SMV coat protein (CP) sequences from seven provinces in Korea using RT-PCR and Sanger sequencing. Phylogenetic and haplotype analyses revealed eight groups of 83 SMV isolates and a network of 50 SMV haplotypes in Korea. The phylogenetic tree using 305 SMV CP sequences available worldwide revealed 12 clades that were further divided into two groups according to the plant hosts. Recombination rarely occurred in the CP sequences, while negative selection was dominant in the SMV CP sequences. Genetic diversity analyses revealed that plant species had a greater impact on the genetic diversity of SMV CP sequences than geographical origin or location. SMV isolates identified from Pinellia species in China showed the highest genetic diversity. Phylodynamic analysis showed that the SMV isolates between the two Pinellia species diverged in the year 1248. Since the divergence of the first SMV isolate from Glycine max in 1486, major clades for SMV isolates infecting Glycine species seem to have diverged from 1791 to 1886. Taken together, we provide a comprehensive overview of the genetic diversity and divergence of SMV CP sequences.
Keywords: coat protein; haplotype; population; soybean mosaic virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Bowers G., Jr., Goodman R. Soybean mosaic virus: Infection of soybean seed parts and seed transmission. Phytopathology. 1979;69:569–572. doi: 10.1094/Phyto-69-569. - DOI
-
- Hartman G.L., West E.D., Herman T.K. Crops that feed the World 2. Soybean—Worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 2011;3:5–17. doi: 10.1007/s12571-010-0108-x. - DOI
-
- Shin D., Jeong D. Korean traditional fermented soybean products: Jang. J. Ethn. Foods. 2015;2:2–7. doi: 10.1016/j.jef.2015.02.002. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

