Quantitative Real-Time PCR Based on SYBR Green Technology for the Identification of Philaenus italosignus Drosopoulos & Remane (Hemiptera Aphrophoridae)
- PMID: 36501353
- PMCID: PMC9741283
- DOI: 10.3390/plants11233314
Quantitative Real-Time PCR Based on SYBR Green Technology for the Identification of Philaenus italosignus Drosopoulos & Remane (Hemiptera Aphrophoridae)
Abstract
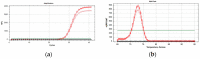
The use of molecular tools to identify insect pests is a critical issue, especially when rapid and reliable tests are required. We proposed a protocol based on qPCR with SYBR Green technology to identify Philaenus italosignus (Hemiptera, Aphrophoridae). The species is one of the three spittlebugs able to transmit Xylella fastidiosa subsp. pauca ST53 in Italy, together with Philaenus spumarius and Neophilaenus campestris. Although less common than the other two species, its identification is key to verifying which role it can play when locally abundant. The proposed assay shows analytical specificity being inclusive with different populations of the target species and exclusive with non-target taxa, either taxonomically related or not. Moreover, it shows analytical sensibility, repeatability, and reproducibility, resulting in an excellent candidate for an official diagnostic method. The molecular test can discriminate P. italosignus from all non-target species, including the congeneric P. spumarius.
Keywords: Xylella fastidiosa vector; alpha taxonomic identification; molecular diagnostic tool; spittlebug.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Drosopoulos S., Remane R. Biogeographic studies on the spittlebug Philaenus signatus Melichar, 1896 species group (Hemiptera: Aphrophoridae) with the description of two new allopatric species. Ann. Soc. Entomol. Fr. N.S. 2000;36:269–277.
-
- Panzavolta T., Bracalini M., Croci F., Ghelardini L., Luti S., Campigli S., Goti E., Marchi R., Tiberi R., Marchi G. Philaenus italosignus a potential vector of Xylella fastidiosa: Occurrence of the spittlebug on olive trees in Tuscany (Italy) Bull. Insectol. 2019;72:317–320.
-
- Marchi G., Rizzo D., Ranaldi F., Ghelardini L., Ricciolini M., Scarpelli I., Drosera L., Goti E., Capretti P., Surico G. First detection of Xylella fastidiosa subsp. multiplex DNA in Tuscany (Italy) Phytopathol. Mediterr. 2018;57:363–364. doi: 10.14601/Phytopathol_Mediterr-24454. - DOI
-
- Saponari M., D’Attoma G., Kubaa R.A., Loconsole G., Altamura G., Zicca S., Rizzo D., Boscia D. A new variant of Xylella fastidiosa subspecies multiplex detected in different host plants in the recently emerged outbreak in the region of Tuscany, Italy. Eur. J. Plant Pathol. 2019;154:1195–1200. doi: 10.1007/s10658-019-01736-9. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

