How to survive pig farming: Mechanism of SCC mec element deletion and metabolic stress adaptation in livestock-associated MRSA
- PMID: 36504815
- PMCID: PMC9728531
- DOI: 10.3389/fmicb.2022.969961
How to survive pig farming: Mechanism of SCC mec element deletion and metabolic stress adaptation in livestock-associated MRSA
Abstract
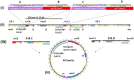
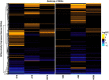
Previous research on methicillin susceptible Staphylococcus aureus (MSSA) belonging to livestock-associated (LA-) sequence type (ST) 398, isolated from pigs and their local surroundings, indicated that differences between these MSSA and their methicillin resistant predecessors (MRSA) are often limited to the absence of the staphylococcal cassette chromosome mec (SCCmec) and few single nucleotide polymorphisms. So far, our understanding on how LA-MRSA endure the environmental conditions associated with pig-farming as well as the putative impact of this particular environment on the mobilisation of SCCmec elements is limited. Thus, we performed in-depth genomic and transcriptomic analyses using the LA-MRSA ST398 strain IMT38951 and its methicillin susceptible descendant. We identified a mosaic-structured SCCmec region including a putative replicative SCCmecVc which is absent from the MSSA chromosome through homologous recombination. Based on our data, such events occur between short repetitive sequences identified within and adjacent to two distinct alleles of the large cassette recombinase genes C (ccrC). We further evaluated the global transcriptomic response of MRSA ST398 to particular pig-farm associated conditions, i.e., contact with host proteins (porcine serum) and a high ammonia concentration. Differential expression of global regulators involved in stress response control were identified, i.e., ammonia-induced alternative sigma factor B-depending activation of genes for the alkaline shock protein 23, the heat shock response and the accessory gene regulator (agr)-controlled transcription of virulence factors. Exposure to serum transiently induced the transcription of distinct virulence factor encoding genes. Transcription of genes reported for mediating the loss of methicillin resistance, especially ccrC, was not significantly different compared to the unchallenged controls. We concluded that, from an evolutionary perspective, bacteria may save energy by incidentally dismissing a fully replicative SCCmec element in contrast to the induction of ccr genes on a population scale. Since the genomic SCCmec integration site is a hot-spot of recombination, occasional losses of elements of 16 kb size may restore capacities for the uptake of foreign genetic material. Subsequent spread of resistance, on the other hand, might depend on the autonomous replication machinery of the deleted SCCmec elements that probably enhance chances for reintegration of SCCmec into susceptible genomes by mere multiplication.
Keywords: SCCmec; ammonium; deletion; livestock associated; manure (litter); methicillin resistant Staphylococcus aureus; recombination; transcriptome analysis.
Copyright © 2022 Huber, Wolf, Ziebuhr, Holmes, Assmann, Lübke-Becker, Thürmer, Semmler, Brombach, Bethe, Bischoff, Wieler, Epping and Walther.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Anderson K. L., Roux C. M., Olson M. W., Luong T. T., Lee C. Y., Olson R., et al. . (2010). Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 60, 208–250. doi: 10.1111/j.1574-695X.2010.00736.x, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

