The roles of KLHL family members in human cancers
- PMID: 36504893
- PMCID: PMC9729911
The roles of KLHL family members in human cancers
Abstract
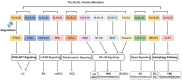
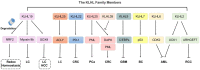
The Kelch-like (KLHL) family members consist of three domains: bric-a-brac, tramtrack, broad complex/poxvirus and zinc finger domain, BACK domain and Kelch domain, which combine and interact with Cullin3 to form an E3 ubiquitin ligase. Research has indicated that KLHL family members ubiquitinate target substrates to regulate physiological and pathological processes, including tumorigenesis and progression. KLHL19, a member of the KLHL family, is associated with tumorigenesis and drug resistance. However, the regulation and cross talks of other KLHL family members, which also play roles in cancer, are still unclear. Our review mainly explores studies concerning the roles of other KLHL family members in tumor-related regulation to provide novel insights into KLHL family members.
Keywords: E3 ubiquitin ligase; Kelch-like family member; degradation; human cancer; ubiquitination.
AJCR Copyright © 2022.
Conflict of interest statement
None.
Figures

Similar articles
-
The dual nature of KLHL proteins: From cellular regulators to disease drivers.Eur J Cell Biol. 2025 Jun;104(2):151483. doi: 10.1016/j.ejcb.2025.151483. Epub 2025 Mar 14. Eur J Cell Biol. 2025. PMID: 40101609 Review.
-
Kelch-like proteins: Physiological functions and relationships with diseases.Pharmacol Res. 2019 Oct;148:104404. doi: 10.1016/j.phrs.2019.104404. Epub 2019 Aug 20. Pharmacol Res. 2019. PMID: 31442578 Review.
-
Update on the Kelch-like (KLHL) gene family.Hum Genomics. 2013 May 15;7(1):13. doi: 10.1186/1479-7364-7-13. Hum Genomics. 2013. PMID: 23676014 Free PMC article.
-
A novel zebrafish kelchlike gene klhl and its human ortholog KLHL display conserved expression patterns in skeletal and cardiac muscles.Gene. 2004 Aug 18;338(1):75-83. doi: 10.1016/j.gene.2004.05.016. Gene. 2004. PMID: 15302408
-
Kelch-like proteins in the gastrointestinal tumors.Acta Pharmacol Sin. 2023 May;44(5):931-939. doi: 10.1038/s41401-022-01007-0. Epub 2022 Oct 20. Acta Pharmacol Sin. 2023. PMID: 36266566 Free PMC article. Review.
Cited by
-
The E3 ubiquitin ligase adaptor KLHL8 targets ZAR1 to regulate maternal mRNA degradation in oocytes.EMBO Rep. 2025 Jul 28. doi: 10.1038/s44319-025-00537-y. Online ahead of print. EMBO Rep. 2025. PMID: 40721554
-
Identifying Genes Associated with the Anticancer Activity of a Fluorinated Chalcone in Triple-Negative Breast Cancer Cells Using Bioinformatics Tools.Int J Mol Sci. 2025 Apr 12;26(8):3662. doi: 10.3390/ijms26083662. Int J Mol Sci. 2025. PMID: 40332279 Free PMC article.
-
Intestinal KLHL12 is dispensable for lipid absorption and chylomicron metabolism.Am J Physiol Endocrinol Metab. 2025 Aug 1;329(2):E233-E240. doi: 10.1152/ajpendo.00219.2025. Epub 2025 Jul 1. Am J Physiol Endocrinol Metab. 2025. PMID: 40591354 Free PMC article.
-
KLHL14 is a tumor suppressor downregulated in undifferentiated thyroid cancer.Cell Death Discov. 2024 Jun 22;10(1):297. doi: 10.1038/s41420-024-02063-7. Cell Death Discov. 2024. PMID: 38909024 Free PMC article.
-
Allelic effects on KLHL17 expression likely mediated by JunB/D underlie a PDAC GWAS signal at chr1p36.33.medRxiv [Preprint]. 2024 Sep 16:2024.09.16.24313748. doi: 10.1101/2024.09.16.24313748. medRxiv. 2024. Update in: Nat Commun. 2025 Apr 30;16(1):4055. doi: 10.1038/s41467-025-59109-2. PMID: 39371158 Free PMC article. Updated. Preprint.
References
Publication types
LinkOut - more resources
Full Text Sources
