Elevated bile acid metabolism and microbiome are associated with suppressed cell proliferation and better survival in breast cancer
- PMID: 36504906
- PMCID: PMC9729904
Elevated bile acid metabolism and microbiome are associated with suppressed cell proliferation and better survival in breast cancer
Abstract
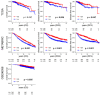
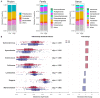
Bile acids are metabolized by the gut microbiome and are involved in fat absorption. Contrary to their carcinogenic role in gastrointestinal cancers, bile acids have been reported to inhibit cancer cell proliferation in breast cancer. The microbiome of breast cancer tissues may also influence cancer proliferation. We hypothesized that bile acid metabolism reflects its accumulation and is associated with certain microbiomes, breast cancer biology, and patient survival. Transcriptomic and clinicopathological information of a total of 6050 patients in three large open primary breast cancer cohorts (GSE96058, METABRIC, TCGA) and 16S rRNA gene sequence microbiome data of breast cancer tissues in TCGA were analyzed by high and low bile acid metabolism scores calculated by gene set variation analysis (GSVA). Breast cancers with high bile acid metabolism had a significantly improved survival across all three cohorts. Metabolic pathways related to the production and regulation of bile acids were consistently enriched in high bile acid metabolism groups across all cohorts. On the other hand, the low bile acid metabolism group was associated with higher Ki67 expression and Nottingham histological grade, as well as enrichment of cell proliferation-related gene sets. Intratumoral heterogeneity, homologous recombination deficiency, mutational load, activation of cancer immunity, and infiltration of anticancer immune cells were also higher in this group. Gammaretrovirus, Hymenobacter, Anaerococcus, and Collimonas were significantly more abundant in the high bile acid metabolism group compared to Lactobacillus, Ruegeria, and Marichromatium in the low metabolism group. Surprisingly, almost all Hallmark cell proliferation-associated gene sets were highly enriched in all three microorganisms that were abundant in the low bile acid metabolism group. In conclusion, microorganisms abundant in the breast tumor microenvironment with low bile acid metabolism are associated with aggressive cancer biology, including increased cell proliferation and poor survival.
Keywords: BRCA; Bile acid; GSVA; breast cancer; microbiome; tumor microenvironment.
AJCR Copyright © 2022.
Conflict of interest statement
None.
Figures




References
-
- Dieleman S, Aarnoutse R, Ziemons J, Kooreman L, Boleij A, Smidt M. Exploring the potential of breast microbiota as biomarker for breast cancer and therapeutic response. Am J Pathol. 2021;191:968–982. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
