Emerging roles of alternative RNA splicing in oral squamous cell carcinoma
- PMID: 36505770
- PMCID: PMC9732560
- DOI: 10.3389/fonc.2022.1019750
Emerging roles of alternative RNA splicing in oral squamous cell carcinoma
Abstract
Alternative RNA splicing (ARS) is an essential and tightly regulated cellular process of post-transcriptional regulation of pre-mRNA. It produces multiple isoforms and may encode proteins with different or even opposite functions. The dysregulated ARS of pre-mRNA contributes to the development of many cancer types, including oral squamous cell carcinoma (OSCC), and may serve as a biomarker for the diagnosis and prognosis of OSCC and an attractive therapeutic target. ARS is mainly regulated by splicing factors, whose expression is also often dysregulated in OSCC and involved in tumorigenesis. This review focuses on the expression and roles of splicing factors in OSCC, the alternative RNA splicing events associated with OSCC, and recent advances in therapeutic approaches that target ARS.
Keywords: OSCC; alternative splicing; splicing factor; therapeutic targets; tumor progression.
Copyright © 2022 Liu, Guo and Jia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
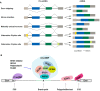
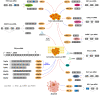
Similar articles
-
Identification of RNA-splicing factor Lsm12 as a novel tumor-associated gene and a potent biomarker in Oral Squamous Cell Carcinoma (OSCC).J Exp Clin Cancer Res. 2022 Apr 21;41(1):150. doi: 10.1186/s13046-022-02355-9. J Exp Clin Cancer Res. 2022. PMID: 35449073 Free PMC article.
-
Identification of a prognostic alternative splicing signature in oral squamous cell carcinoma.J Cell Physiol. 2020 May;235(5):4804-4813. doi: 10.1002/jcp.29357. Epub 2019 Oct 21. J Cell Physiol. 2020. PMID: 31637730
-
SRSF5 functions as a novel oncogenic splicing factor and is upregulated by oncogene SRSF3 in oral squamous cell carcinoma.Biochim Biophys Acta Mol Cell Res. 2018 Sep;1865(9):1161-1172. doi: 10.1016/j.bbamcr.2018.05.017. Epub 2018 May 30. Biochim Biophys Acta Mol Cell Res. 2018. PMID: 29857020
-
RNA splicing: a dual-edged sword for hepatocellular carcinoma.Med Oncol. 2022 Aug 16;39(11):173. doi: 10.1007/s12032-022-01726-8. Med Oncol. 2022. PMID: 35972700 Review.
-
Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
References
Publication types
LinkOut - more resources
Full Text Sources

