FK506-binding protein, FKBP12, promotes serine utilization and negatively regulates threonine deaminase in fission yeast
- PMID: 36505930
- PMCID: PMC9730122
- DOI: 10.1016/j.isci.2022.105659
FK506-binding protein, FKBP12, promotes serine utilization and negatively regulates threonine deaminase in fission yeast
Abstract
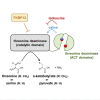
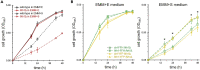
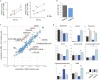
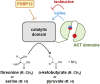
FK506-binding protein with a molecular weight of 12 kDa (FKBP12) is a receptor of the immunosuppressive drugs, FK506 and rapamycin. The physiological functions of FKBP12 remain ambiguous because of its nonessentiality and multifunctionality. Here, we show that FKBP12 promotes the utilization of serine as a nitrogen source and regulates the isoleucine biosynthetic pathway in fission yeast. In screening for small molecules that inhibit serine assimilation, we found that the growth of fission yeast cells in medium supplemented with serine as the sole nitrogen source, but not in glutamate-supplemented medium, was suppressed by FKBP12 inhibitors. Knockout of FKBP12 phenocopied the action of these compounds in serine-supplemented medium. Metabolome analyses and genetic screens identified the threonine deaminase, Tda1, to be regulated downstream of FKBP12. Genetic and biochemical analyses unveiled the negative regulation of Tda1 by FKBP12. Our findings reveal new roles of FKBP12 in amino acid biosynthesis and nitrogen metabolism homeostasis.
Keywords: Biochemistry; Biological sciences; Biosynthesis; Cell biology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
FK506-binding protein (FKBP12) regulates ryanodine receptor-evoked Ca2+ release in colonic but not aortic smooth muscle.Cell Calcium. 2008 Jun;43(6):539-49. doi: 10.1016/j.ceca.2007.09.002. Epub 2007 Oct 24. Cell Calcium. 2008. PMID: 17950843
-
FK506 binding protein 12 mediates sensitivity to both FK506 and rapamycin in murine mast cells.Eur J Immunol. 1995 Feb;25(2):563-71. doi: 10.1002/eji.1830250239. Eur J Immunol. 1995. PMID: 7533090
-
Physiological function of FKBP12, a primary target of rapamycin/FK506: a newly identified role in transcription of ribosomal protein genes in yeast.Curr Genet. 2021 Jun;67(3):383-388. doi: 10.1007/s00294-020-01142-3. Epub 2021 Jan 12. Curr Genet. 2021. PMID: 33438053 Review.
-
Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin.Plant J. 1998 Aug;15(4):511-9. doi: 10.1046/j.1365-313x.1998.00232.x. Plant J. 1998. PMID: 9753776
-
Interactions of FK506 and Rapamycin With FK506 Binding Protein 12 in Opportunistic Human Fungal Pathogens.Front Mol Biosci. 2020 Oct 16;7:588913. doi: 10.3389/fmolb.2020.588913. eCollection 2020. Front Mol Biosci. 2020. PMID: 33195437 Free PMC article. Review.
Cited by
-
Identification and characterization of ternary complexes consisting of FKBP12, MAPRE1 and macrocyclic molecular glues.RSC Chem Biol. 2025 Mar 6;6(5):788-799. doi: 10.1039/d4cb00279b. eCollection 2025 May 8. RSC Chem Biol. 2025. PMID: 40059881 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

