Regulation of hierarchical carbon substrate utilization, nitrogen fixation, and root colonization by the Hfq/Crc/CrcZY genes in Pseudomonas stutzeri
- PMID: 36505936
- PMCID: PMC9730152
- DOI: 10.1016/j.isci.2022.105663
Regulation of hierarchical carbon substrate utilization, nitrogen fixation, and root colonization by the Hfq/Crc/CrcZY genes in Pseudomonas stutzeri
Abstract
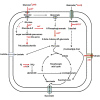
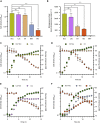
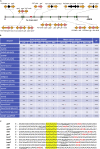
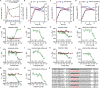
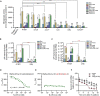
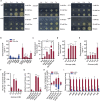
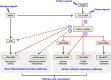
Bacteria of the genus Pseudomonas consume preferred carbon substrates in nearly reverse order to that of enterobacteria, and this process is controlled by RNA-binding translational repressors and regulatory ncRNA antagonists. However, their roles in microbe-plant interactions and the underlying mechanisms remain uncertain. Here we show that root-associated diazotrophic Pseudomonas stutzeri A1501 preferentially catabolizes succinate, followed by the less favorable substrate citrate, and ultimately glucose. Furthermore, the Hfq/Crc/CrcZY regulatory system orchestrates this preference and contributes to optimal nitrogenase activity and efficient root colonization. Hfq has a central role in this regulatory network through different mechanisms of action, including repressing the translation of substrate-specific catabolic genes, activating the nitrogenase gene nifH posttranscriptionally, and exerting a positive effect on the transcription of an exopolysaccharide gene cluster. Our results illustrate an Hfq-mediated mechanism linking carbon metabolism to nitrogen fixation and root colonization, which may confer rhizobacteria competitive advantages in rhizosphere environments.
Keywords: Plant genetics; plant biology; plant nutrition.
© 2022.
Conflict of interest statement
The authors have no competing interests to declare.
Figures

References
-
- Görke B., Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. - PubMed
-
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008;11:87–93. - PubMed
-
- Magasanik B. In: The Lactose Operon. Beckwith J., Zipser D., editors. Cold Spring Harbor Laboratory Press; 1970. Glucose effects: inducer exclusion and repression; pp. 189–220.
-
- Silby M.W., Winstanley C., Godfrey S.A.C., Levy S.B., Jackson R.W. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 2011;35:652–680. - PubMed
LinkOut - more resources
Full Text Sources

