A novel diG motif in ORF3a protein of SARS-Cov-2 for intracellular transport
- PMID: 36506095
- PMCID: PMC9727819
- DOI: 10.3389/fcell.2022.1011221
A novel diG motif in ORF3a protein of SARS-Cov-2 for intracellular transport
Abstract
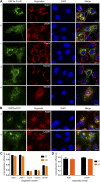
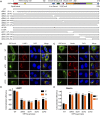
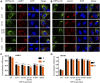
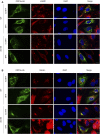
The ongoing SARS-CoV-2/COVID-19 pandemic caused a global public health crisis. Yet, everyone's response to SARS-CoV-2 infection varies, and different viral variants confer diverse pathogenicity. Thus, it is imperative to understand how viral determinants contribute to COVID-19. Viral ORF3a protein is one of those viral determinants, as its functions are linked to induction of cell and tissues damages, disease severity and cytokine storm that is a major cause of COVID-19-related death. ORF3a is a membrane-associated protein. Upon synthesis, it is transported from endoplasmic reticulum, Golgi apparatus to plasma membrane and subcellular endomembranes including endosomes and lysosomes. However, how ORF3a is transported intracellularly remains elusive. The goal of this study was to carry out a systematic mutagenesis study to determine the structural relationship of ORF3a protein with its subcellular locations. Single amino acid (aa) and deletion mutations were generated in the putative function-relevant motifs and other regions of interest. Immunofluorescence and ImageJ analyses were used to determine and quantitate subcellular locations of ORF3a mutants in comparison with wildtype ORF3a. The wildtype ORF3a localizes predominantly (Pearson's coefficients about 0.8) on the membranes of endosomes and lysosomes. Consistent with earlier findings, deletion of the YXXΦ motif, which is required for protein export, retained ORF3a in the Golgi apparatus. Interestingly, mutations in a double glycine (diG) region (aa 187-188) displayed a similar phenotype to the YXXΦ deletion, implicating a similar role of the diG motif in intracellular transport. Indeed, interrupting any one of the two glycine residues such as deletion of a single (dG188), both (dG187/dG188) or substitution (G188Y) of these residues led to ORF3a retention in the Golgi apparatus (Pearson's coefficients ≥0.8). Structural analyses further suggest that the diG motif supports a type-II β-turn between the anti-parallel β4 and β5 sheets and connects to the YXXΦ motif via hydrogen bonds between two monomers. The diG- YXXΦ interaction forms a hand-in-hand configuration that could facilitate dimerization. Together, these observations suggest a functional role of the diG motif in intracellular transport of ORF3a.
Keywords: Golgi apparatus; ORF3a; SARS-CoV-2; diG motif; diG-YXXΦ interaction; intracellular transport; lysosome; mutagenesis.
Copyright © 2022 Cruz-Cosme, Zhang, Liu, Mahase, Sallapalli, Chang, Zhang, Teng, Zhao and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Targeting the YXXΦ Motifs of the SARS Coronaviruses 1 and 2 ORF3a Peptides by In Silico Analysis to Predict Novel Virus-Host Interactions.Biomolecules. 2022 Jul 29;12(8):1052. doi: 10.3390/biom12081052. Biomolecules. 2022. PMID: 36008946 Free PMC article.
-
Deletion of both the Tyrosine-Based Endocytosis Signal and the Endoplasmic Reticulum Retrieval Signal in the Cytoplasmic Tail of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Pigs.J Virol. 2019 Jan 4;93(2):e01758-18. doi: 10.1128/JVI.01758-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30404797 Free PMC article.
-
The YXXΦ motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport.Virol J. 2014 Apr 24;11:75. doi: 10.1186/1743-422X-11-75. Virol J. 2014. PMID: 24762043 Free PMC article.
-
Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19.Front Microbiol. 2022 Mar 9;13:854567. doi: 10.3389/fmicb.2022.854567. eCollection 2022. Front Microbiol. 2022. PMID: 35356515 Free PMC article. Review.
-
Regulation of autophagy by SARS-CoV-2: The multifunctional contributions of ORF3a.J Med Virol. 2023 Jul;95(7):e28959. doi: 10.1002/jmv.28959. J Med Virol. 2023. PMID: 37485696 Review.
Cited by
-
Advanced Protocol for Molecular Characterization of Viral Genome in Fission Yeast (Schizosaccharomyces pombe).Pathogens. 2024 Jul 4;13(7):566. doi: 10.3390/pathogens13070566. Pathogens. 2024. PMID: 39057793 Free PMC article.
-
Activity and cellular distribution of ORF3a mutants of SARS-CoV-2 variants of concern.J Gen Virol. 2025 Aug;106(8):002135. doi: 10.1099/jgv.0.002135. J Gen Virol. 2025. PMID: 40768270 Free PMC article.
-
Subcellular localization of SARS-CoV-2 E and 3a proteins along the secretory pathway.J Mol Histol. 2025 Mar 1;56(2):98. doi: 10.1007/s10735-025-10375-w. J Mol Histol. 2025. PMID: 40025386 Free PMC article.
-
SARS-CoV-2 ORF3a-Mediated NF-κB Activation Is Not Dependent on TRAF-Binding Sequence.Viruses. 2023 Nov 8;15(11):2229. doi: 10.3390/v15112229. Viruses. 2023. PMID: 38005906 Free PMC article.
-
SARS-CoV-2 ORF3a induces COVID-19-associated kidney injury through HMGB1-mediated cytokine production.mBio. 2024 Nov 13;15(11):e0230824. doi: 10.1128/mbio.02308-24. Epub 2024 Sep 30. mBio. 2024. PMID: 39345136 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

