Developing methylotrophic microbial platforms for a methanol-based bioindustry
- PMID: 36507257
- PMCID: PMC9727194
- DOI: 10.3389/fbioe.2022.1050740
Developing methylotrophic microbial platforms for a methanol-based bioindustry
Abstract
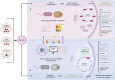
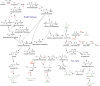
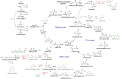
Methanol, a relatively cheap and renewable single-carbon feedstock, has gained considerable attention as a substrate for the bio-production of commodity chemicals. Conventionally produced from syngas, along with emerging possibilities of generation from methane and CO2, this C1 substrate can serve as a pool for sequestering greenhouse gases while supporting a sustainable bio-economy. Methylotrophic organisms, with the inherent ability to use methanol as the sole carbon and energy source, are competent candidates as platform organisms. Accordingly, methanol bioconversion pathways have been an attractive target for biotechnological and bioengineering interventions in developing microbial cell factories. This review summarizes the recent advances in methanol-based production of various bulk and value-added chemicals exploiting the native and synthetic methylotrophic organisms. Finally, the current challenges and prospects of streamlining these methylotrophic platforms are discussed.
Keywords: bio-based chemical production; metabolic engineering; methanol; methylotrophs; microbial cell factories; synthetic biology.
Copyright © 2022 Singh, Kang, Kwon and Kim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Anthony C. (1982). The biochemistry of methylotrophs. Cambridge: Academic Press.
-
- Beekwilder J., van Houwelingen A., Cankar K., van Dijk A. D., de Jong R. M., Stoopen G., et al. (2014). Valencene synthase from the heartwood of N ootka cypress (C allitropsis nootkatensis) for biotechnological production of valencene. Plant Biotechnol. J. 12 (2), 174–182. 10.1111/pbi.12124 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

