Genomic Characterization of Skin and Soft Tissue Streptococcus pyogenes Isolates from a Low-Income and a High-Income Setting
- PMID: 36507654
- PMCID: PMC9942559
- DOI: 10.1128/msphere.00469-22
Genomic Characterization of Skin and Soft Tissue Streptococcus pyogenes Isolates from a Low-Income and a High-Income Setting
Abstract
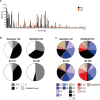
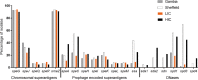
Streptococcus pyogenes is a leading cause of human morbidity and mortality, especially in resource-limited settings. The development of a vaccine against S. pyogenes is a global health priority to reduce the burden of postinfection rheumatic heart disease. To support this, molecular characterization of circulating S. pyogenes isolates is needed. We performed whole-genome analyses of S. pyogenes isolates from skin and soft tissue infections in Sukuta, The Gambia, a low-income country (LIC) in West Africa where there is a high burden of such infections. To act as a comparator to these LIC isolates, skin infection isolates from Sheffield, United Kingdom (a high-income country [HIC]), were also sequenced. The LIC isolates from The Gambia were genetically more diverse (46 emm types in 107 isolates) than the HIC isolates from Sheffield (23 emm types in 142 isolates), with only 7 overlapping emm types. Other molecular markers were shared, including a high prevalence of the skin infection-associated emm pattern D and the variable fibronectin-collagen-T antigen (FCT) types FCT-3 and FCT-4. Fewer of the Gambian LIC isolates carried prophage-associated superantigens (64%) and DNases (26%) than did the Sheffield HIC isolates (99% and 95%, respectively). We also identified streptococcin genes unique to 36% of the Gambian LIC isolates and a higher prevalence (48%) of glucuronic acid utilization pathway genes in the Gambian LIC isolates than in the Sheffield HIC isolates (26%). Comparison to a wider collection of HIC and LIC isolate genomes supported our findings of differing emm diversity and prevalence of bacterial factors. Our study provides insight into the genetics of LIC isolates and how they compare to HIC isolates. IMPORTANCE The global burden of rheumatic heart disease (RHD) has triggered a World Health Organization response to drive forward development of a vaccine against the causative human pathogen Streptococcus pyogenes. This burden stems primarily from low- and middle-income settings where there are high levels of S. pyogenes skin and soft tissue infections, which can lead to RHD. Our study provides much needed whole-genome-based molecular characterization of isolates causing skin infections in Sukuta, The Gambia, a low-income country (LIC) in West Africa where infection and RHD rates are high. Although we identified a greater level of diversity in these LIC isolates than in isolates from Sheffield, United Kingdom (a high-income country), there were some shared features. There were also some features that differed by geographical region, warranting further investigation into their contribution to infection. Our study has also contributed data essential for the development of a vaccine that would target geographically relevant strains.
Keywords: FCT region; bacterial genomics; emm types; group A Streptococcus; skin infection; vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Watkins DA, Johnson CO, Colquhoun SM, Karthikeyan G, Beaton A, Bukhman G, Forouzanfar MH, Longenecker CT, Mayosi BM, Mensah GA, Nascimento BR, Ribeiro ALP, Sable CA, Steer AC, Naghavi M, Mokdad AH, Murray CJL, Vos T, Carapetis JR, Roth GA. 2017. Global, regional, and national burden of rheumatic heart disease, 1990-2015. N Engl J Med 377:713–722. doi: 10.1056/NEJMoa1603693. - DOI - PubMed
-
- Vekemans J, Gouvea-Reis F, Kim JH, Excler J-L, Smeesters PR, O'Brien KL, Van Beneden CA, Steer AC, Carapetis JR, Kaslow DC. 2019. The path to group A Streptococcus vaccines: World Health Organization research and development technology roadmap and preferred product characteristics. Clin Infect Dis 69:877–883. doi: 10.1093/cid/ciy1143. - DOI - PMC - PubMed
-
- Pastural É, McNeil SA, MacKinnon-Cameron D, Ye L, Langley JM, Stewart R, Martin LH, Hurley GJ, Salehi S, Penfound TA, Halperin S, Dale JB. 2020. Safety and immunogenicity of a 30-valent M protein-based group a streptococcal vaccine in healthy adult volunteers: a randomized, controlled phase I study. Vaccine 38:1384–1392. doi: 10.1016/j.vaccine.2019.12.005. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
