Linolenic Acid-Derived Oxylipins Inhibit Aflatoxin Biosynthesis in Aspergillus flavus through Activation of Imizoquin Biosynthesis
- PMID: 36508213
- PMCID: PMC9785051
- DOI: 10.1021/acs.jafc.2c06230
Linolenic Acid-Derived Oxylipins Inhibit Aflatoxin Biosynthesis in Aspergillus flavus through Activation of Imizoquin Biosynthesis
Abstract
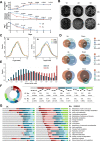
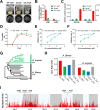
Oxylipins play important signaling roles in aflatoxin (AF) biosynthesis in Aspergillus flavus. We previously showed that exogenous supply of autoxidated linolenic acid (AL) inhibited AF biosynthesis in A. flavus via oxylipins, but the molecular mechanism is still unknown. Here, we performed multiomics analyses of A. flavus grown in media with or without AL. Targeted metabolite analyses and quantitative reverse transcription (qRT)-polymerase chain reaction (PCR) showed that the imizoquin (IMQ) biosynthetic pathway was distinctly upregulated in the presence of AL. 13C-glucose labeling confirmed in parallel that the tricarboxylic acid cycle was also enhanced by AL, consistent with observed increases in mycelial growth. Moreover, we integrated thermal proteome profiling and molecular dynamics simulations to identify a potential receptor of AL; AL was found to interact with a transporter (ImqJ) located in the IMQ gene cluster, primarily through hydrophobic interactions. Further analyses of strains with an IMQ pathway transcription factor overexpressed or knocked out confirmed that this pathway was critical for AL-mediated inhibition of AF biosynthesis. Comparison of 22 assembled A. flavus and Aspergillus oryzae genomes showed that genes involved in the IMQ pathway were positively selected in A. oryzae. Taken together, the results of our study provide novel insights into oxylipin-mediated regulation of AF biosynthesis and suggest potential methods for preventing AF contamination of crops.
Keywords: Aspergillus flavus; aflatoxins; imizoquin; multiomics; thermal proteome profiling.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Liao J.; He Z.; Xia Y.; Lei Y.; Liao B. A review on biosynthesis and genetic regulation of aflatoxin production by major Aspergillus fungi. Oil Crop Sci. 2020, 5, 166–173. 10.1016/j.ocsci.2020.11.001. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

