Microtubule-associated protein 1B is implicated in stem cell commitment and nervous system regeneration in planarians
- PMID: 36508441
- PMCID: PMC9744283
- DOI: 10.1371/journal.pone.0278966
Microtubule-associated protein 1B is implicated in stem cell commitment and nervous system regeneration in planarians
Abstract
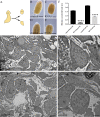
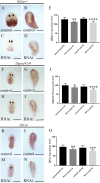
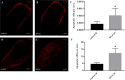
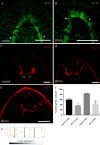
Microtubule-associated 1B (MAP1B) proteins are expressed at the nervous system level where they control cytoskeleton activity and regulate neurotransmitter release. Here, we report about the identification of a planarian MAP1B factor (DjMap1B) that is enriched in cephalic ganglia and longitudinal nerve cords but not in neoblasts, the plentiful population of adult stem cells present in planarians, thanks to which these animals can continuously cell turnover and regenerate any lost body parts. DjMap1B knockdown induces morphological anomalies in the nervous system and affects neoblast commitment. Our data put forward a correlation between a MAP1B factor and stem cells and suggest a function of the nervous system in non-cell autonomous control of planarian stem cells.
Copyright: © 2022 Gambino et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Gambino G, Rossi L & Salvetti A (2022) Planarian stem cells: pluripotency maintenance and fate determination. In Advances in Aquatic Invertebrate Stem Cell Research From Basic Research to Innovative Application, 221–241. MDPI, Basel, Switzerland, doi: 10.3390/books978-3-0365-1635-6-6 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

