Algorithmic identification of atypical diabetes in electronic health record (EHR) systems
- PMID: 36508462
- PMCID: PMC9744270
- DOI: 10.1371/journal.pone.0278759
Algorithmic identification of atypical diabetes in electronic health record (EHR) systems
Abstract
Aims: Understanding atypical forms of diabetes (AD) may advance precision medicine, but methods to identify such patients are needed. We propose an electronic health record (EHR)-based algorithmic approach to identify patients who may have AD, specifically those with insulin-sufficient, non-metabolic diabetes, in order to improve feasibility of identifying these patients through detailed chart review.
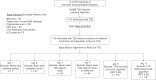
Methods: Patients with likely T2D were selected using a validated machine-learning (ML) algorithm applied to EHR data. "Typical" T2D cases were removed by excluding individuals with obesity, evidence of dyslipidemia, antibody-positive diabetes, or cystic fibrosis. To filter out likely type 1 diabetes (T1D) cases, we applied six additional "branch algorithms," relying on various clinical characteristics, which resulted in six overlapping cohorts. Diabetes type was classified by manual chart review as atypical, not atypical, or indeterminate due to missing information.
Results: Of 114,975 biobank participants, the algorithms collectively identified 119 (0.1%) potential AD cases, of which 16 (0.014%) were confirmed after expert review. The branch algorithm that excluded T1D based on outpatient insulin use had the highest percentage yield of AD (13 of 27; 48.2% yield). Together, the 16 AD cases had significantly lower BMI and higher HDL than either unselected T1D or T2D cases identified by ML algorithms (P<0.05). Compared to the ML T1D group, the AD group had a significantly higher T2D polygenic score (P<0.01) and lower hemoglobin A1c (P<0.01).
Conclusion: Our EHR-based algorithms followed by manual chart review identified collectively 16 individuals with AD, representing 0.22% of biobank enrollees with T2D. With a maximum yield of 48% cases after manual chart review, our algorithms have the potential to drastically improve efficiency of AD identification. Recognizing patients with AD may inform on the heterogeneity of T2D and facilitate enrollment in studies like the Rare and Atypical Diabetes Network (RADIANT).
Copyright: © 2022 Cromer et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: a close family member of SJC is employed by a Johnson & Johnson company. This does not alter our adherence to PLOS ONE policies on sharing data and materials. The other authors report no competing interests.
Figures


References
-
- National Diabetes Statistics Report, 2020 | CDC. 28 Sep 2020 [cited 18 Sep 2021]. Available: https://www.cdc.gov/diabetes/data/statistics-report/index.html
-
- 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021 | Diabetes Care | American Diabetes Association. [cited 9 Feb 2022]. Available: https://diabetesjournals.org/care/article/44/Supplement_1/S15/30859/2-Cl... - PubMed
-
- NIH funds first nationwide network to study rare forms of diabetes. In: National Institutes of Health (NIH) [Internet]. 30 Sep 2020 [cited 9 Feb 2022]. Available: https://www.nih.gov/news-events/news-releases/nih-funds-first-nationwide...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

