Case-control investigation of invasive Salmonella disease in Malawi reveals no evidence of environmental or animal transmission of invasive strains, and supports human to human transmission
- PMID: 36508466
- PMCID: PMC9779717
- DOI: 10.1371/journal.pntd.0010982
Case-control investigation of invasive Salmonella disease in Malawi reveals no evidence of environmental or animal transmission of invasive strains, and supports human to human transmission
Abstract
Background: Invasive Salmonella infections cause significant morbidity and mortality in Sub-Saharan Africa. However, the routes of transmission are uncertain. We conducted a case-control study of index-case and geographically-matched control households in Blantyre, Malawi, sampling Salmonella isolates from index cases, healthy people, animals, and the household environment.
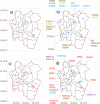
Methodology: Sixty index cases of human invasive Salmonella infection were recruited (March 2015-Oct 2016). Twenty-eight invasive Non-Typhoidal Salmonella (iNTS) disease and 32 typhoid patients consented to household sampling. Each index-case household was geographically matched to a control household. Extensive microbiological sampling included stool sampling from healthy household members, stool or rectal swabs from household-associated animals and boot-sock sampling of the household environment.
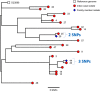
Findings: 1203 samples from 120 households, yielded 43 non-Typhoidal Salmonella (NTS) isolates from 25 households (overall sample positivity 3.6%). In the 28 iNTS patients, disease was caused by 3 STs of Salmonella Typhimurium, mainly ST313. In contrast, the isolates from households spanned 15 sequence types (STs). Two S. Typhimurium isolates from index cases closely matched isolates from their respective asymptomatic household members (2 and 3 SNP differences respectively). Despite the recovery of a diverse range of NTS, there was no overlap between the STs causing iNTS disease with any environmental or animal isolates.
Conclusions: The finding of NTS strains from index cases that matched household members, coupled with lack of related animal or environmental isolates, supports a hypothesis of human to human transmission of iNTS infections in the household. The breadth of NTS strains found in animals and the household environment demonstrated the robustness of NTS sampling and culture methodology, and suggests a diverse ecology of Salmonella in this setting. Healthy typhoid (S. Typhi) carrier state was not detected. The lack of S. Typhi isolates from the household environment suggests that further methodological development is needed to culture S. Typhi from the environment.
Copyright: © 2022 Koolman et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Supporting evidence for a human reservoir of invasive non-Typhoidal Salmonella from household samples in Burkina Faso.PLoS Negl Trop Dis. 2019 Oct 14;13(10):e0007782. doi: 10.1371/journal.pntd.0007782. eCollection 2019 Oct. PLoS Negl Trop Dis. 2019. PMID: 31609964 Free PMC article.
-
Intestinal carriage of invasive non-typhoidal Salmonella among household members of children with Salmonella bloodstream infection, Kisangani, DR Congo.Front Microbiol. 2023 Oct 12;14:1241961. doi: 10.3389/fmicb.2023.1241961. eCollection 2023. Front Microbiol. 2023. PMID: 37901802 Free PMC article.
-
Incidence of invasive salmonella disease in sub-Saharan Africa: a multicentre population-based surveillance study.Lancet Glob Health. 2017 Mar;5(3):e310-e323. doi: 10.1016/S2214-109X(17)30022-0. Lancet Glob Health. 2017. PMID: 28193398 Free PMC article.
-
Antimicrobial resistance and management of invasive Salmonella disease.Vaccine. 2015 Jun 19;33 Suppl 3(0 3):C21-9. doi: 10.1016/j.vaccine.2015.03.102. Epub 2015 Apr 23. Vaccine. 2015. PMID: 25912288 Free PMC article. Review.
-
Epidemiology and Genomics of Invasive Nontyphoidal Salmonella Infections in Kenya.Clin Infect Dis. 2015 Nov 1;61 Suppl 4(Suppl 4):S317-24. doi: 10.1093/cid/civ711. Clin Infect Dis. 2015. PMID: 26449947 Free PMC article. Review.
Cited by
-
Outbreak detection in Harar town and Kersa district, Ethiopia using phylogenetic analysis and source attribution.BMC Infect Dis. 2024 Aug 26;24(1):864. doi: 10.1186/s12879-024-09800-4. BMC Infect Dis. 2024. PMID: 39187763 Free PMC article.
-
Genomic investigation and nationwide tracking of pediatric invasive nontyphoidal Salmonella in China.mLife. 2024 Mar 29;3(1):156-160. doi: 10.1002/mlf2.12117. eCollection 2024 Mar. mLife. 2024. PMID: 38827503 Free PMC article.
-
Immunogenicity and protective efficacy of nanoparticle formulations of L-SseB against Salmonella infection.Front Immunol. 2023 Jun 30;14:1208848. doi: 10.3389/fimmu.2023.1208848. eCollection 2023. Front Immunol. 2023. PMID: 37457702 Free PMC article.
-
Salmonella enterica serovar Typhimurium ST313 sublineage 2.2 has emerged in Malawi with a characteristic gene expression signature and a fitness advantage.Microlife. 2024 Mar 28;5:uqae005. doi: 10.1093/femsml/uqae005. eCollection 2024. Microlife. 2024. PMID: 38623411 Free PMC article.
-
Landscape analysis of invasive non-typhoidal salmonella (iNTS) disease and iNTS vaccine use case and demand: Report of a WHO expert consultation.Vaccine. 2025 May 10;55:127008. doi: 10.1016/j.vaccine.2025.127008. Epub 2025 Mar 24. Vaccine. 2025. PMID: 40132323 Free PMC article.
References
-
- Stanaway JD, Reiner RC, Blacker BF, Goldberg EM, Khalil IA, Troeger CE, et al.. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet Infectious Diseases. 2019;19: 369–381. doi: 10.1016/S1473-3099(18)30685-6 - DOI - PMC - PubMed
-
- Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, et al.. The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet Infectious Diseases. 2019;19: 1312–1324. doi: 10.1016/S1473-3099(19)30418-9 - DOI - PMC - PubMed
-
- Hendriksen RS, Leekitcharoenphon P, Lukjancenko O, Lukwesa-Musyani C, Tambatamba B, Mwaba J, et al.. Genomic Signature of Multidrug-Resistant Salmonella enterica Serovar Typhi Isolates Related to a Massive Outbreak in Zambia between 2010 and 2012. Munson E, editor. J Clin Microbiol. 2015;53: 262–272. doi: 10.1128/JCM.02026-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

