Comparison between relative and absolute quantitative real-time PCR applied to single-cell analyses: Transcriptional levels in a key neuron for long-term memory in the pond snail
- PMID: 36508476
- PMCID: PMC9744327
- DOI: 10.1371/journal.pone.0279017
Comparison between relative and absolute quantitative real-time PCR applied to single-cell analyses: Transcriptional levels in a key neuron for long-term memory in the pond snail
Abstract
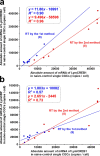
Quantitative real-time PCR (qPCR) is a powerful method for measuring nucleic acid levels and quantifying mRNA levels, even in single cells. In the present study, we compared the results of single-cell qPCR obtained by different quantification methods (relative and absolute) and different reverse transcription methods. In the experiments, we focused on the cerebral giant cell (CGC), a key neuron required for the acquisition of conditioned taste aversion in the pond snail Lymnaea stagnalis, and examined changes in the mRNA levels of 3 memory-related genes, cAMP-response element binding proteins (LymCREB1 and LymCREB2) and CREB-binding protein (LymCBP), during memory formation. The results obtained by relative quantification showed similar patterns for the 3 genes. For absolute quantification, reverse transcription was performed using 2 different methods: a mixture of oligo d(T) primers and random primers (RT method 1); and gene-specific primers (RT method 2). These methods yielded different results and did not show consistent changes related to conditioning. The mRNA levels in the samples prepared by RT method 2 were up to 3.3 times higher than those in samples prepared by RT method 1. These results suggest that for qPCR of single neurons, the efficacy and validity do not differ between relative and absolute quantification methods, but the reverse transcription step critically influences the results of mRNA quantification.
Copyright: © 2022 Hatakeyama et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

