Erythroid/megakaryocytic differentiation confers BCL-XL dependency and venetoclax resistance in acute myeloid leukemia
- PMID: 36508699
- PMCID: PMC10651789
- DOI: 10.1182/blood.2021011094
Erythroid/megakaryocytic differentiation confers BCL-XL dependency and venetoclax resistance in acute myeloid leukemia
Abstract
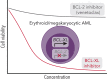
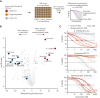
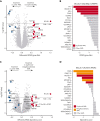
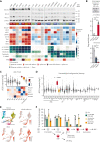
Myeloid neoplasms with erythroid or megakaryocytic differentiation include pure erythroid leukemia, myelodysplastic syndrome with erythroid features, and acute megakaryoblastic leukemia (FAB M7) and are characterized by poor prognosis and limited treatment options. Here, we investigate the drug sensitivity landscape of these rare malignancies. We show that acute myeloid leukemia (AML) cells with erythroid or megakaryocytic differentiation depend on the antiapoptotic protein B-cell lymphoma (BCL)-XL, rather than BCL-2, using combined ex vivo drug sensitivity testing, genetic perturbation, and transcriptomic profiling. High-throughput screening of >500 compounds identified the BCL-XL-selective inhibitor A-1331852 and navitoclax as highly effective against erythroid/megakaryoblastic leukemia cell lines. In contrast, these AML subtypes were resistant to the BCL-2 inhibitor venetoclax, which is used clinically in the treatment of AML. Consistently, genome-scale CRISPR-Cas9 and RNAi screening data demonstrated the striking essentiality of BCL-XL-encoding BCL2L1 but not BCL2 or MCL1, for the survival of erythroid/megakaryoblastic leukemia cell lines. Single-cell and bulk transcriptomics of patient samples with erythroid and megakaryoblastic leukemias identified high BCL2L1 expression compared with other subtypes of AML and other hematological malignancies, where BCL2 and MCL1 were more prominent. BCL-XL inhibition effectively killed blasts in samples from patients with AML with erythroid or megakaryocytic differentiation ex vivo and reduced tumor burden in a mouse erythroleukemia xenograft model. Combining the BCL-XL inhibitor with the JAK inhibitor ruxolitinib showed synergistic and durable responses in cell lines. Our results suggest targeting BCL-XL as a potential therapy option in erythroid/megakaryoblastic leukemias and highlight an AML subgroup with potentially reduced sensitivity to venetoclax-based treatments.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: H.K. reports research funding from AbbVie and personal fees from Faron outside the submitted work. U.W.-K. has received honoraria from Sanofi, Novartis, and Pfizer (not related to this study) and is a member of the advisory boards of Gilead, Pfizer, and Jazz Pharmaceuticals. C.A.H. has received research funding from Celgene, Kronos Bio, Novartis, Oncopeptides, Orion, and IMI2 projects HARMONY and HARMONY PLUS unrelated to this work. S.M. has received research funding and honoraria from BMS, Novartis, and Pfizer (not related to this study). The remaining authors declare no competing financial interests.
Figures



Comment in
-
Is BCL-xL the Achilles' heel of AEL and AMKL?Blood. 2023 Mar 30;141(13):1505-1506. doi: 10.1182/blood.2022019246. Blood. 2023. PMID: 36995706 No abstract available.
References
-
- Wickrema A, Crispino JD. Erythroid and megakaryocytic transformation. Oncogene. 2007;26(47):6803–6815. - PubMed
-
- Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–2302. - PubMed
-
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

