Metabolites enhance innate resistance to human Mycobacterium tuberculosis infection
- PMID: 36509283
- PMCID: PMC9746823
- DOI: 10.1172/jci.insight.152357
Metabolites enhance innate resistance to human Mycobacterium tuberculosis infection
Abstract
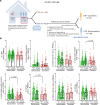
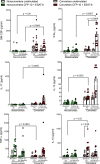
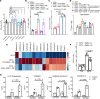
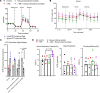
To determine the mechanisms that mediate resistance to Mycobacterium tuberculosis (M. tuberculosis) infection in household contacts (HHCs) of patients with tuberculosis (TB), we followed 452 latent TB infection-negative (LTBI-) HHCs for 2 years. Those who remained LTBI- throughout the study were identified as nonconverters. At baseline, nonconverters had a higher percentage of CD14+ and CD3-CD56+CD27+CCR7+ memory-like natural killer (NK) cells. Using a whole-transcriptome and metabolomic approach, we identified deoxycorticosterone acetate as a metabolite with elevated concentrations in the plasma of nonconverters, and further studies showed that this metabolite enhanced glycolytic ATP flux in macrophages and restricted M. tuberculosis growth by enhancing antimicrobial peptide production through the expression of the surface receptor sialic acid binding Ig-like lectin-14. Another metabolite, 4-hydroxypyridine, from the plasma of nonconverters significantly enhanced the expansion of memory-like NK cells. Our findings demonstrate that increased levels of specific metabolites can regulate innate resistance against M. tuberculosis infection in HHCs of patients with TB who never develop LTBI or active TB.
Keywords: Immunology; Tuberculosis.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

