p21 induces a senescence program and skeletal muscle dysfunction
- PMID: 36509362
- PMCID: PMC9800630
- DOI: 10.1016/j.molmet.2022.101652
p21 induces a senescence program and skeletal muscle dysfunction
Abstract
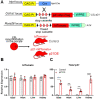
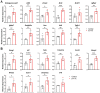
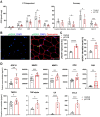
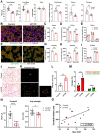
Recent work has established associations between elevated p21, the accumulation of senescent cells, and skeletal muscle dysfunction in mice and humans. Using a mouse model of p21 overexpression (p21OE), we examined if p21 mechanistically contributes to cellular senescence and pathological features in skeletal muscle. We show that p21 induces several core properties of cellular senescence in skeletal muscle, including an altered transcriptome, DNA damage, mitochondrial dysfunction, and the senescence-associated secretory phenotype (SASP). Furthermore, p21OE mice exhibit manifestations of skeletal muscle pathology, such as atrophy, fibrosis, and impaired physical function when compared to age-matched controls. These findings suggest p21 alone is sufficient to drive a cellular senescence program and reveal a novel source of skeletal muscle loss and dysfunction.
Keywords: Aging; Cellular senescence; DNA damage; Fibrosis; Physical function; Sarcopenia; Senescence-associated secretory phenotype.
Copyright © 2022 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

