Clinical strains of Mycobacterium tuberculosis exhibit differential lipid metabolism-associated transcriptome changes in in vitro cholesterol and infection models
- PMID: 36509392
- PMCID: PMC9936260
- DOI: 10.1093/femspd/ftac046
Clinical strains of Mycobacterium tuberculosis exhibit differential lipid metabolism-associated transcriptome changes in in vitro cholesterol and infection models
Abstract
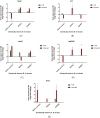
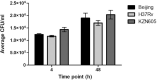
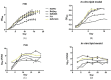
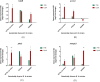
Many studies have identified host-derived lipids, characterised by the abundance of cholesterol, as a major source of carbon nutrition for Mycobacterium tuberculosis during infection. Members of the Mycobacterium tuberculosis complex are biologically different with regards to degree of disease, host range, pathogenicity and transmission. Therefore, the current study aimed at elucidating transcriptome changes during early infection of pulmonary epithelial cells and on an in vitro cholesterol-rich minimal media, in M. tuberculosis clinical strains F15/LAM4/KZN and Beijing, and the laboratory H37Rv strain. Infection of pulmonary epithelial cells elicited the upregulation of fadD28 and hsaC in both the F15/LAM4/KZN and Beijing strains and the downregulation of several other lipid-associated genes. Growth curve analysis revealed F15/LAM4/KZN and Beijing to be slow growers in 7H9 medium and cholesterol-supplemented media. RNA-seq analysis revealed strain-specific transcriptomic changes, thereby affecting different metabolic processes in an in vitro cholesterol model. The differential expression of these genes suggests that the genetically diverse M. tuberculosis clinical strains exhibit strain-specific behaviour that may influence their ability to metabolise lipids, specifically cholesterol, which may account for phenotypic differences observed during infection.
Keywords: Mycobacterium tuberculosis; cholesterol; clinical strains; lipid metabolism; transcriptome.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

