Targeting PEA3 transcription factors to mitigate small cell lung cancer progression
- PMID: 36509998
- PMCID: PMC9898033
- DOI: 10.1038/s41388-022-02558-6
Targeting PEA3 transcription factors to mitigate small cell lung cancer progression
Abstract
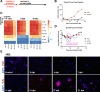
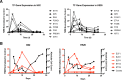
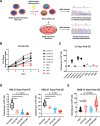
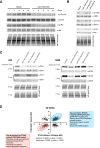
Small cell lung cancer (SCLC) remains a lethal disease with a dismal overall survival rate of 6% despite promising responses to upfront combination chemotherapy. The key drivers of such rapid mortality include early metastatic dissemination in the natural course of the disease and the near guaranteed emergence of chemoresistant disease. Here, we found that we could model the regression and relapse seen in clinical SCLC in vitro. We utilized time-course resolved RNA-sequencing to globally profile transcriptome changes as SCLC cells responded to a combination of cisplatin and etoposide-the standard-of-care in SCLC. Comparisons across time points demonstrated a distinct transient transcriptional state resembling embryonic diapause. Differential gene expression analysis revealed that expression of the PEA3 transcription factors ETV4 and ETV5 were transiently upregulated in the surviving fraction of cells which we determined to be necessary for efficient clonogenic expansion following chemotherapy. The FGFR-PEA3 signaling axis guided the identification of a pan-FGFR inhibitor demonstrating in vitro and in vivo efficacy in delaying progression following combination chemotherapy, observed inhibition of phosphorylation of the FGFR adaptor FRS2 and corresponding downstream MAPK and PI3K-Akt signaling pathways. Taken together, these data nominate PEA3 transcription factors as key mediators of relapse progression in SCLC and identify a clinically actionable small molecule candidate for delaying relapse of SCLC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17:765. - PubMed
-
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

