Characterization of two 1,3-β-glucan-modifying enzymes from Penicillium sumatraense reveals new insights into 1,3-β-glucan metabolism of fungal saprotrophs
- PMID: 36510318
- PMCID: PMC9745967
- DOI: 10.1186/s13068-022-02233-8
Characterization of two 1,3-β-glucan-modifying enzymes from Penicillium sumatraense reveals new insights into 1,3-β-glucan metabolism of fungal saprotrophs
Abstract
Background: 1,3-β-glucan is a polysaccharide widely distributed in the cell wall of several phylogenetically distant organisms, such as bacteria, fungi, plants and microalgae. The presence of highly active 1,3-β-glucanases in fungi evokes the biological question on how these organisms can efficiently metabolize exogenous sources of 1,3-β-glucan without incurring in autolysis.
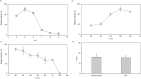
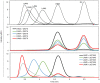
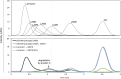
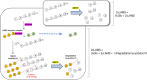
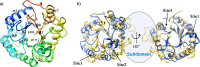
Results: To elucidate the molecular mechanisms at the basis of 1,3-β-glucan metabolism in fungal saprotrophs, the putative exo-1,3-β-glucanase G9376 and a truncated form of the putative glucan endo-1,3-β-glucosidase (ΔG7048) from Penicillium sumatraense AQ67100 were heterologously expressed in Pichia pastoris and characterized both in terms of activity and structure. G9376 efficiently converted laminarin and 1,3-β-glucan oligomers into glucose by acting as an exo-glycosidase, whereas G7048 displayed a 1,3-β-transglucanase/branching activity toward 1,3-β-glucan oligomers with a degree of polymerization higher than 5, making these oligomers more recalcitrant to the hydrolysis acted by exo-1,3-β-glucanase G9376. The X-ray crystallographic structure of the catalytic domain of G7048, solved at 1.9 Å of resolution, consists of a (β/α)8 TIM-barrel fold characteristic of all the GH17 family members. The catalytic site is in a V-shaped cleft containing the two conserved catalytic glutamic residues. Molecular features compatible with the activity of G7048 as 1,3-β-transglucanase are discussed.
Conclusions: The antagonizing activity between ΔG7048 and G9376 indicates how opportunistic fungi belonging to Penicillium genus can feed on substrates similar for composition and structure to their own cell wall without incurring in a self-deleterious autohydrolysis.
Keywords: 1,3-β-Glucan metabolism; 1,3-β-Transglucanase; Cell wall-modifying enzymes; Exo-1,3-β-glucanase; Penicillium; TIM-barrel.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Purification, characterization and synergism in autolysis of a group of 1,3-β-glucan hydrolases from the pilei of Coprinopsis cinerea fruiting bodies.Microbiology (Reading). 2015 Oct;161(10):1978-1989. doi: 10.1099/mic.0.000143. Epub 2015 Jul 21. Microbiology (Reading). 2015. PMID: 26199012
-
Derepression of beta-1,3-glucanases in Penicillium italicum: localization of the various enzymes and correlation with cell wall glucan mobilization and autolysis.J Bacteriol. 1979 Jan;137(1):6-12. doi: 10.1128/jb.137.1.6-12.1979. J Bacteriol. 1979. PMID: 762023 Free PMC article.
-
Effect of beta-glucanases on Penicillium oxalicum cell wall fractions.FEMS Microbiol Lett. 1990 Aug;58(3):233-9. doi: 10.1111/j.1574-6968.1990.tb13984.x. FEMS Microbiol Lett. 1990. PMID: 2121589
-
Microbial β-glucanases: production, properties, and engineering.World J Microbiol Biotechnol. 2023 Feb 27;39(4):106. doi: 10.1007/s11274-023-03550-2. World J Microbiol Biotechnol. 2023. PMID: 36847914 Review.
-
α-1,3-Glucanase: present situation and prospect of research.World J Microbiol Biotechnol. 2016 Feb;32(2):30. doi: 10.1007/s11274-015-1977-0. Epub 2016 Jan 9. World J Microbiol Biotechnol. 2016. PMID: 26748807 Review.
Cited by
-
Research advances in fungal polysaccharides: production, extraction, characterization, properties, and their multifaceted applications.Front Cell Infect Microbiol. 2025 Jun 9;15:1604184. doi: 10.3389/fcimb.2025.1604184. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40552121 Free PMC article. Review.
-
A First Expression, Purification and Characterization of Endo-β-1,3-Glucanase from Penicillium expansum.J Fungi (Basel). 2023 Sep 25;9(10):961. doi: 10.3390/jof9100961. J Fungi (Basel). 2023. PMID: 37888217 Free PMC article.
-
Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2021-2022.Mass Spectrom Rev. 2025 May-Jun;44(3):213-453. doi: 10.1002/mas.21873. Epub 2024 Jun 24. Mass Spectrom Rev. 2025. PMID: 38925550 Free PMC article. Review.
References
-
- Lange L, Agger JW, Meyer AS. Fungal Biotechnology: Unlocking the full potential of fungi for a more sustainable world. In: Grand Challenge in Fungal Biotechnology. 2020; 1:3–32.
-
- Benedetti M, Locci F, Gramegna G, Sestili F, Savarin DV. Green production and biotechnological applications of cell wall lytic enzymes. Appl. Sci. 2019;9(23):5012. doi: 10.3390/app9235012. - DOI
-
- Giovannoni M, Larini I, Scafati V, Scortica A, Compri M, Pontiggia D, Zapparoli G, Vitulo N, Benedetti M, Mattei B. A novel Penicillium sumatraense isolate reveals an arsenal of degrading enzymes exploitable in algal bio-refinery processes. Biotechnol Biofuels. 2021;14(1):20. doi: 10.1186/s13068-021-02030-9. - DOI - PMC - PubMed
-
- Mendoza-Mendoza A, Zaid R, Lawry R, Hermosa R, Monte E, Horwitz AB, Mukherjee PK. Molecular dialogues between Trichoderma and roots: Role of the fungal secretome. Fungal Biology Reviews. 2018;32:62–85. doi: 10.1016/j.fbr.2017.12.001. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
