Dual Domestication, Diversity, and Differential Introgression in Old World Cotton Diploids
- PMID: 36510772
- PMCID: PMC9792962
- DOI: 10.1093/gbe/evac170
Dual Domestication, Diversity, and Differential Introgression in Old World Cotton Diploids
Abstract
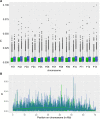
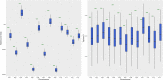
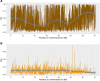
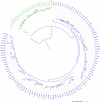
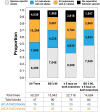
Domestication in the cotton genus is remarkable in that it has occurred independently four different times at two different ploidy levels. Relatively little is known about genome evolution and domestication in the cultivated diploid species Gossypium herbaceum and Gossypium arboreum, due to the absence of wild representatives for the latter species, their ancient domestication, and their joint history of human-mediated dispersal and interspecific gene flow. Using in-depth resequencing of a broad sampling from both species, we provide support for their independent domestication, as opposed to a progenitor-derivative relationship, showing that diversity (mean π = 6 × 10-3) within species is similar, and that divergence between species is modest (FST = 0.413). Individual accessions were homozygous for ancestral single-nucleotide polymorphisms at over half of variable sites, while fixed, derived sites were at modest frequencies. Notably, two chromosomes with a paucity of fixed, derived sites (i.e., chromosomes 7 and 10) were also strongly implicated as having experienced high levels of introgression. Collectively, these data demonstrate variable permeability to introgression among chromosomes, which we propose is due to divergent selection under domestication and/or the phenomenon of F2 breakdown in interspecific crosses. Our analyses provide insight into the evolutionary forces that shape diversity and divergence in the diploid cultivated species and establish a foundation for understanding the contribution of introgression and/or strong parallel selection to the extensive morphological similarities shared between species.
Keywords: Gossypium arboreum; Gossypium herbaceum; cotton; domestication; introgression.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- R Core Team . 2020. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.
-
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320–2325. - PubMed
-
- Applequist WL, Cronn R, Wendel JF. 2001. Comparative development of fiber in wild and cultivated cotton. Evol Dev. 3:3–17. - PubMed
-
- Basu AK. 1996. Current genetic research in cotton in India. Genetica. 97:279–290.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

