Characteristics of the ErmK Protein of Bacillus halodurans C-125
- PMID: 36511701
- PMCID: PMC9927578
- DOI: 10.1128/spectrum.02598-22
Characteristics of the ErmK Protein of Bacillus halodurans C-125
Abstract
Bacillus halodurans C-125 is an alkaliphilic microorganism that grows best at pH 10 to 10.5. B. halodurans C-125 harbors the erm (erythromycin resistance methylase) gene as well as the mphB (macrolide phosphotransferase) and putative mef (macrolide efflux) genes, which confer resistance to macrolide, lincosamide, and streptogramin B (MLSB) antibiotics. The Erm protein expressed in B. halodurans C-125 could be classified as ErmK because it shares 66.2% and 61.2% amino acid sequence identity with the closest ErmD and Erm(34), respectively. ErmK can be regarded as a dimethylase, as evidenced by reverse transcriptase analysis and the antibiotic resistance profile exhibited by E. coli expressing ermK. Although ErmK showed one-third or less in vitro methylating activity compared to ErmC', E. coli cells expressing ErmK exhibited comparable resistance to erythromycin and tylosin, and a similar dimethylation proportion of 23S rRNA due to the higher expression rate in a T7 promoter-mediated expression system. The less efficient methylation activity of ErmK might reflect an adaption to mitigate the fitness cost caused by dimethylation through the Erm protein presumably because B. halodurans C-125 has less probability to encounter the antibiotics in its favorable growth conditions and grows retardedly in neutral environments. IMPORTANCE Erm proteins confer MLSB antibiotic resistance (minimal inhibitory concentration [MIC] value up to 4,096 μg/mL) on microorganisms ranging from antibiotic producers to pathogens, imposing one of the most pressing threats to clinics. Therefore, Erm proteins have long been speculated to be plausible targets for developing inhibitor(s). In our laboratory, it has been noticed that there are variations in enzymatic activity among the Erm proteins, Erm in antibiotic producers being better than that in pathogens. In this study, it has been observed that Erm protein in B. halodurans C-125 extremophile is a novel member of Erm protein and acts more laggardly, compared to that in pathogen. While this sluggishness of Erm protein in extremophile might be evolved to reduce the fitness cost incurred by Erm activity adapting to its environments, this feature could be exploited to develop the more potent and/or efficacious drug to combat formidably problematic antibiotic-resistant pathogens.
Keywords: antibiotic resistance; extromophile; fitness cost; methylases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
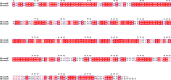
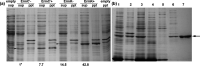


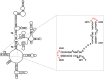
References
-
- Honda H, Kudo T, Ikura Y, Horikoshi K. 1985. Two types of xylanases of alkalophilic Bacillus sp. No. C-125. Can J Microbiol 31:538–542. doi:10.1139/m85-100. - DOI
-
- Ikura Y, Horikoshi K. 1979. Isolation and some properties of β-Galactosidase producing bacteria. Agric Biol Chem 43:85–88. doi:10.1080/00021369.1979.10863410. - DOI
-
- Patel S, Gupta RS. 2020. A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int J Syst Evol Micribiol 70:406–438. doi:10.1099/ijsem.0.003775. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

