Proteomics Profiling Reveals Regulation of Immune Response to Salmonella enterica Serovar Typhimurium Infection in Mice
- PMID: 36511704
- PMCID: PMC9872662
- DOI: 10.1128/iai.00499-22
Proteomics Profiling Reveals Regulation of Immune Response to Salmonella enterica Serovar Typhimurium Infection in Mice
Abstract
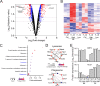
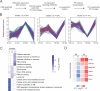
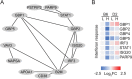
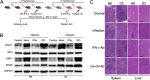
Regulation of the immune response to Salmonella enterica serovar Typhimurium (S. Typhimurium) infection is a complex process, influenced by the interaction between genetic and environmental factors. Different inbred strains of mice exhibit distinct levels of resistance to S. Typhimurium infection, ranging from susceptible (e.g., C57BL/6J) to resistant (e.g., DBA/2J) strains. However, the underlying molecular mechanisms contributing to the host response remain elusive. In this study, we present a comprehensive proteomics profiling of spleen tissue from C57BL/6J and DBA/2J strains with different doses of S. Typhimurium infection by tandem mass tag labeling coupled with two-dimensional liquid chromatography-tandem mass spectrometry (TMT-LC/LC-MS/MS). We identified and quantified 3,986 proteins, resulting in 475 differentially expressed proteins (DEPs) between C57BL/6J and DBA/2J strains. Functional enrichment analysis unveiled that the mechanisms of innate immune responses to S. Typhimurium infection could be associated with several signaling pathways, including the interferon (IFN) signaling pathway. We experimentally validated the roles of the IFN signaling pathway in the innate immune response to S. Typhimurium infection using an IFN-γ neutralization assay. We further illustrated the importance of macrophage and proinflammatory cytokines in the mechanisms underlying the resistance to S. Typhimurium using quantitative reverse transcription-PCR (qRT-PCR). Taken together, our results provided new insights into the genetic regulation of the immune response to S. Typhimurium infection in mice and might lead to the discovery of potential protein targets for controlling salmonellosis.
Keywords: S. Typhimurium; Salmonella; genetic regulation; immune response; mass spectrometry; mouse; proteome; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Differential immune responses of C57BL/6 mice to infection by Salmonella enterica serovar Typhimurium strain SL1344, CVCC541 and CMCC50115.Virulence. 2019 Dec;10(1):248-259. doi: 10.1080/21505594.2019.1597496. Virulence. 2019. PMID: 30898022 Free PMC article.
-
An attenuated Salmonella enterica serovar Typhimurium strain lacking the ZnuABC transporter induces protection in a mouse intestinal model of Salmonella infection.Vaccine. 2011 Feb 17;29(9):1783-90. doi: 10.1016/j.vaccine.2010.12.111. Epub 2011 Jan 8. Vaccine. 2011. PMID: 21219981
-
Genomic Analysis of Salmonella enterica Serovar Typhimurium Characterizes Strain Diversity for Recent U.S. Salmonellosis Cases and Identifies Mutations Linked to Loss of Fitness under Nitrosative and Oxidative Stress.mBio. 2016 Mar 8;7(2):e00154. doi: 10.1128/mBio.00154-16. mBio. 2016. PMID: 26956590 Free PMC article.
-
Innate immune control of Salmonella enterica serovar Typhimurium: mechanisms contributing to combating systemic Salmonella infection.J Innate Immun. 2011;3(6):543-9. doi: 10.1159/000330771. Epub 2011 Sep 10. J Innate Immun. 2011. PMID: 21912097 Review.
-
Immunity to salmonellosis.Immunol Rev. 2011 Mar;240(1):196-210. doi: 10.1111/j.1600-065X.2010.00999.x. Immunol Rev. 2011. PMID: 21349095 Review.
References
-
- Centers for Disease Control and Prevention. 2022. Salmonella. https://www.cdc.gov/salmonella/index.html.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

