Single-cell RNA sequencing reveals distinct T cell populations in immune-related adverse events of checkpoint inhibitors
- PMID: 36513074
- PMCID: PMC9873824
- DOI: 10.1016/j.xcrm.2022.100868
Single-cell RNA sequencing reveals distinct T cell populations in immune-related adverse events of checkpoint inhibitors
Abstract
PD-1 is an inhibitory receptor in T cells, and antibodies that block its interaction with ligands augment anti-tumor immune responses. The clinical potential of these agents is limited by the fact that half of all patients develop immune-related adverse events (irAEs). To generate insights into the cellular changes that occur during anti-PD-1 treatment, we performed single-cell RNA sequencing of circulating T cells collected from patients with cancer. Using the K-nearest-neighbor-based network graph-drawing layout, we show the involvement of distinctive genes and subpopulations of T cells. We identify that at baseline, patients with arthritis have fewer CD8 TCM cells, patients with pneumonitis have more CD4 TH2 cells, and patients with thyroiditis have more CD4 TH17 cells when compared with patients who do not develop irAEs. These data support the hypothesis that different populations of T cells are associated with different irAEs and that characterization of these cells' pre-treatment has the potential to serve as a toxicity-specific predictive biomarker.
Keywords: PD-1; T cells; arthritis; autoimmunity; checkpoint inhibitor; immune-related adverse events; irAEs; pneumonitis; single-cell RNA sequencing; thyroiditis.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

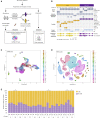




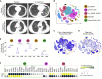
Comment in
-
Distinct T cell sub-clusters may serve as biomarkers for immune related adverse events.Cell Rep Med. 2023 Jan 17;4(1):100902. doi: 10.1016/j.xcrm.2022.100902. Cell Rep Med. 2023. PMID: 36652911 Free PMC article.
References
-
- Rizvi N.A., Mazières J., Planchard D., Stinchcombe T.E., Dy G.K., Antonia S.J., Horn L., Lena H., Minenza E., Mennecier B., et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16:257–265. doi: 10.1016/S1470-2045(15)70054-9. - DOI - PMC - PubMed
-
- Weber J.S., D'Angelo S.P., Minor D., Hodi F.S., Gutzmer R., Neyns B., Hoeller C., Khushalani N.I., Miller W.H., Jr., Lao C.D., et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–384. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

