Molecular characterization and genetic authentication assay for Anopheles 'hemocyte-like' cell lines 4a-3A and 4a-3B
- PMID: 36514125
- PMCID: PMC9749150
- DOI: 10.1186/s13071-022-05590-3
Molecular characterization and genetic authentication assay for Anopheles 'hemocyte-like' cell lines 4a-3A and 4a-3B
Abstract
Background: Anopheles cell lines are used in a variety of ways to better understand the major vectors of malaria in sub-Saharan Africa. Despite this, commonly used cell lines are not well characterized, and no tools are available for cell line identification and authentication.
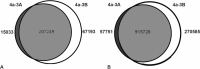
Methods: Utilizing whole genome sequencing, genomes of 4a-3A and 4a-3B 'hemocyte-like' cell lines were characterized for insertions and deletions (indels) and SNP variation. Genomic locations of distinguishing sequence variation and species origin of the cell lines were also examined. Unique indels were targeted to develop a PCR-based cell line authentication assay. Mitotic chromosomes were examined to survey the cytogenetic landscape for chromosome structure and copy number in the cell lines.
Results: The 4a-3A and 4a-3B cell lines are female in origin and primarily of Anopheles coluzzii ancestry. Cytogenetic analysis indicates that the two cell lines are essentially diploid, with some relatively minor chromosome structural rearrangements. Whole-genome sequence was generated, and analysis indicated that SNPs and indels which differentiate the cell lines are clustered on the 2R chromosome in the regions of the 2Rb, 2Rc and 2Ru chromosomal inversions. A PCR-based authentication assay was developed to fingerprint three indels unique to each cell line. The assay distinguishes between 4a-3A and 4a-3B cells and also uniquely identifies two additional An. coluzzii cell lines tested, Ag55 and Sua4.0. The assay has the specificity to distinguish four cell lines and also has the sensitivity to detect cellular contamination within a sample of cultured cells.
Conclusions: Genomic characterization of the 4a-3A and 4a-3B Anopheles cell lines was used to develop a simple diagnostic assay that can distinguish these cell lines within and across research laboratories. A cytogenetic survey indicated that the 4a-3A and Sua4.0 cell lines carry essentially normal diploid chromosomes, which makes them amenable to CRISPR/Cas9 genome editing. The presented simple authentication assay, coupled with screening for mycoplasma, will allow validation of the integrity of experimental resources and will promote greater experimental reproducibility of results.
Keywords: Anopheles coluzzii; Anopheles gambiae; Cell line authentication; Hemocytes; Reproducibility.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Riaz MA, Adang MJ, Hua G, Rezende TMT, Rezende AM, Shen GM. Identification of Lysinibacillus sphaericus binary toxin binding proteins in a malarial mosquito cell line by proteomics: a novel approach towards improving mosquito control. J Proteomics. 2020;227:103918. doi: 10.1016/j.jprot.2020.103918. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

