Cell-type-specific alternative splicing in the Arabidopsis germline
- PMID: 36515615
- PMCID: PMC10152659
- DOI: 10.1093/plphys/kiac574
Cell-type-specific alternative splicing in the Arabidopsis germline
Abstract
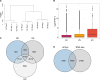
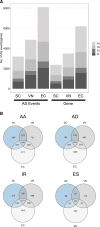
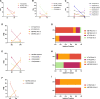
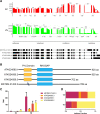
During sexual reproduction in flowering plants, the two haploid sperm cells (SCs) embedded within the cytoplasm of a growing pollen tube are carried to the embryo sac for double fertilization. Pollen development in flowering plants is a dynamic process that encompasses changes at transcriptome and epigenome levels. While the transcriptome of pollen and SCs in Arabidopsis (Arabidopsis thaliana) is well documented, previous analyses have mostly been based on gene-level expression. In-depth transcriptome analysis, particularly the extent of alternative splicing (AS) at the resolution of SC and vegetative nucleus (VN), is still lacking. Therefore, we performed RNA-seq analysis to generate a spliceome map of Arabidopsis SCs and VN isolated from mature pollen grains. Based on our de novo transcriptome assembly, we identified 58,039 transcripts, including 9,681 novel transcripts, of which 2,091 were expressed in SCs and 3,600 in VN. Four hundred and sixty-eight genes were regulated both at gene and splicing levels, with many having functions in mRNA splicing, chromatin modification, and protein localization. Moreover, a comparison with egg cell RNA-seq data uncovered sex-specific regulation of transcription and splicing factors. Our study provides insights into a gamete-specific AS landscape at unprecedented resolution.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




References
-
- Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou X, Russell SD, Sundaresan V (2013) Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76(5): 729–741 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

