Human CARMIL2 deficiency underlies a broader immunological and clinical phenotype than CD28 deficiency
- PMID: 36515678
- PMCID: PMC9754768
- DOI: 10.1084/jem.20220275
Human CARMIL2 deficiency underlies a broader immunological and clinical phenotype than CD28 deficiency
Abstract
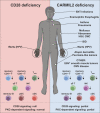
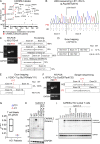
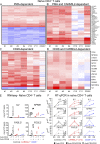
Patients with inherited CARMIL2 or CD28 deficiency have defective T cell CD28 signaling, but their immunological and clinical phenotypes remain largely unknown. We show that only one of three CARMIL2 isoforms is produced and functional across leukocyte subsets. Tested mutant CARMIL2 alleles from 89 patients and 52 families impair canonical NF-κB but not AP-1 and NFAT activation in T cells stimulated via CD28. Like CD28-deficient patients, CARMIL2-deficient patients display recalcitrant warts and low blood counts of CD4+ and CD8+ memory T cells and CD4+ TREGs. Unlike CD28-deficient patients, they have low counts of NK cells and memory B cells, and their antibody responses are weak. CARMIL2 deficiency is fully penetrant by the age of 10 yr and is characterized by numerous infections, EBV+ smooth muscle tumors, and mucocutaneous inflammation, including inflammatory bowel disease. Patients with somatic reversions of a mutant allele in CD4+ T cells have milder phenotypes. Our study suggests that CARMIL2 governs immunological pathways beyond CD28.
© 2022 Lévy et al.
Conflict of interest statement
Disclosures: J. Raedler reported grants from Foerderprogramm fuer Forschung und Lehre (FöFoLe), Medical Faculty, LMU Munich, Germany outside the submitted work. A. Fasth reported personal fees from Lipum AB (rheumatology, advisory board) outside the submitted work. S. Baris reported grants from TUBITAK outside the submitted work. A. Kumar reported personal fees from SOBI and SpringWorks Therapeutics outside the submitted work. No other disclosures were reported.
Figures











References
-
- Alazami, A.M., Al-Helale M., Alhissi S., Al-Saud B., Alajlan H., Monies D., Shah Z., Abouelhoda M., Arnaout R., Al-Dhekri H., et al. . 2018. Novel CARMIL2 Mutations in Patients with Variable Clinical Dermatitis, Infections, and Combined Immunodeficiency. Front. Immunol. 9(203). 10.3389/fimmu.2018.00203 - DOI - PMC - PubMed
-
- Atschekzei, F., Jacobs R., Wetzke M., Sogkas G., Schröder C., Ahrenstorf G., Dhingra A., Ott H., Baumann U., and Schmidt R.E.. 2019. A Novel CARMIL2 Mutation Resulting in Combined Immunodeficiency Manifesting with Dermatitis, Fungal, and Viral Skin Infections As Well as Selective Antibody Deficiency. J. Clin. Immunol. 39:274–276. 10.1007/s10875-019-00628-1 - DOI - PubMed
-
- Béziat, V., Rapaport F., Hu J., Titeux M., Bonnet des Claustres M., Bourgey M., Griffin H., Bandet É., Ma C.S., Sherkat R., et al. . 2021. Humans with inherited T cell CD28 deficiency are susceptible to skin papillomaviruses but are otherwise healthy. Cell. 184:3812–3828.e30. 10.1016/j.cell.2021.06.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

