The injured sciatic nerve atlas (iSNAT), insights into the cellular and molecular basis of neural tissue degeneration and regeneration
- PMID: 36515985
- PMCID: PMC9829412
- DOI: 10.7554/eLife.80881
The injured sciatic nerve atlas (iSNAT), insights into the cellular and molecular basis of neural tissue degeneration and regeneration
Abstract
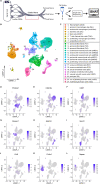
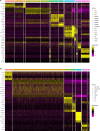
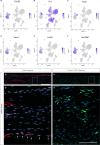
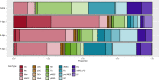
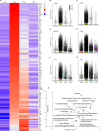
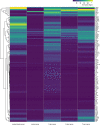
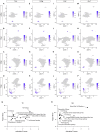
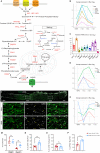
Upon trauma, the adult murine peripheral nervous system (PNS) displays a remarkable degree of spontaneous anatomical and functional regeneration. To explore extrinsic mechanisms of neural repair, we carried out single-cell analysis of naïve mouse sciatic nerve, peripheral blood mononuclear cells, and crushed sciatic nerves at 1 day, 3 days, and 7 days following injury. During the first week, monocytes and macrophages (Mo/Mac) rapidly accumulate in the injured nerve and undergo extensive metabolic reprogramming. Proinflammatory Mo/Mac with a high glycolytic flux dominate the early injury response and rapidly give way to inflammation resolving Mac, programmed toward oxidative phosphorylation. Nerve crush injury causes partial leakiness of the blood-nerve barrier, proliferation of endoneurial and perineurial stromal cells, and entry of opsonizing serum proteins. Micro-dissection of the nerve injury site and distal nerve, followed by single-cell RNA-sequencing, identified distinct immune compartments, triggered by mechanical nerve wounding and Wallerian degeneration, respectively. This finding was independently confirmed with Sarm1-/- mice, in which Wallerian degeneration is greatly delayed. Experiments with chimeric mice showed that wildtype immune cells readily enter the injury site in Sarm1-/- mice, but are sparse in the distal nerve, except for Mo. We used CellChat to explore intercellular communications in the naïve and injured PNS and report on hundreds of ligand-receptor interactions. Our longitudinal analysis represents a new resource for neural tissue regeneration, reveals location- specific immune microenvironments, and reports on large intercellular communication networks. To facilitate mining of scRNAseq datasets, we generated the injured sciatic nerve atlas (iSNAT): https://cdb-rshiny.med.umich.edu/Giger_iSNAT/.
Keywords: Wallerian degeneration; immunology; inflammation; mouse; neuroscience; parabiosis; regeneration; sciatic nerve; single-cell RNA seq.
© 2022, Zhao, Huffman, Hafner et al.
Conflict of interest statement
XZ, LH, HH, MA, MF, AK, RK, CF, RP, CJ, DK, RK, LY, JT, DG, RG No competing interests declared, GC Except for Gabriel Corfas, the authors declare no competing financial or non-financial interests. Gabriel Corfas is a scientific founder of Decibel Therapeutics; he has an equity interest in and has received compensation for consulting. The company was not involved in this study
Figures



















References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

