AKTIP loss is enriched in ERα-positive breast cancer for tumorigenesis and confers endocrine resistance
- PMID: 36516775
- PMCID: PMC9837615
- DOI: 10.1016/j.celrep.2022.111821
AKTIP loss is enriched in ERα-positive breast cancer for tumorigenesis and confers endocrine resistance
Abstract
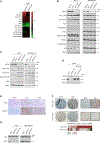
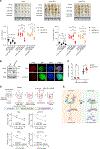
Recurrent deletion of 16q12.2 is observed in luminal breast cancer, yet the causal genomic alterations in this region are largely unknown. In this study, we identify that loss of AKTIP, which is located on 16q12.2, drives tumorigenesis of estrogen receptor alpha (ERα)-positive, but not ERα-negative, breast cancer cells and is associated with poor prognosis of patients with ERα-positive breast cancer. Intriguingly, AKTIP-depleted tumors have increased ERα protein level and activity. Cullin-associated and neddylation-dissociated protein 1 (CAND1), which regulates the cullin-RING E3 ubiquitin ligases, protects ERα from cullin 2-dependent proteasomal degradation. Apart from ERα signaling, AKTIP loss triggers JAK2-STAT3 activation, which provides an alternative survival signal when ERα is inhibited. AKTIP-depleted MCF7 cells and ERα-positive patient-derived organoids are more resistant to ERα antagonists. Importantly, the resistance can be overcome by co-inhibition of JAK2/STAT3. Together, our results highlight the subtype-specific functional consequences of AKTIP loss and provide a mechanistic explanation for the enriched AKTIP copy-number loss in ERα-positive breast cancer.
Keywords: CP: Cancer; endocrine resistance; estrogen receptor; luminal breast cancer; protein degradation.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Chia SK, Bramwell VH, Tu D, Shepherd LE, Jiang S, Vickery T, Mardis E, Leung S, Ung K, Pritchard KI, et al. (2012). A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin. Cancer Res. 18, 4465–4472. 10.1158/1078-0432.CCR-12-0286. - DOI - PMC - PubMed
-
- Ellis MJ, and Perou CM (2013). The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov. 3, 27–34. 10.1158/2159-8290.CD-12-0462. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

