Recurrent repeat expansions in human cancer genomes
- PMID: 36517591
- PMCID: PMC9812771
- DOI: 10.1038/s41586-022-05515-1
Recurrent repeat expansions in human cancer genomes
Abstract
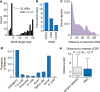
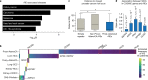
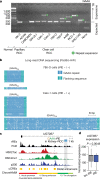
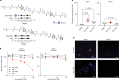
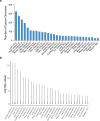
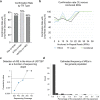
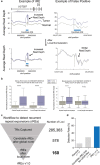
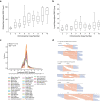
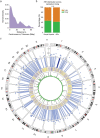
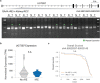
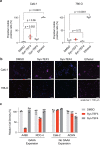
Expansion of a single repetitive DNA sequence, termed a tandem repeat (TR), is known to cause more than 50 diseases1,2. However, repeat expansions are often not explored beyond neurological and neurodegenerative disorders. In some cancers, mutations accumulate in short tracts of TRs, a phenomenon termed microsatellite instability; however, larger repeat expansions have not been systematically analysed in cancer3-8. Here we identified TR expansions in 2,622 cancer genomes spanning 29 cancer types. In seven cancer types, we found 160 recurrent repeat expansions (rREs), most of which (155/160) were subtype specific. We found that rREs were non-uniformly distributed in the genome with enrichment near candidate cis-regulatory elements, suggesting a potential role in gene regulation. One rRE, a GAAA-repeat expansion, located near a regulatory element in the first intron of UGT2B7 was detected in 34% of renal cell carcinoma samples and was validated by long-read DNA sequencing. Moreover, in preliminary experiments, treating cells that harbour this rRE with a GAAA-targeting molecule led to a dose-dependent decrease in cell proliferation. Overall, our results suggest that rREs may be an important but unexplored source of genetic variation in human cancer, and we provide a comprehensive catalogue for further study.
© 2022. The Author(s).
Conflict of interest statement
G.S.E. and M.P.S. are inventors on a patent application describing anti-proliferative agents. E.D. and M.A.E. are shareholders and are currently or were formerly employed by Illumina and Pacific Biosciences. The other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

