A microbiome-dependent gut-brain pathway regulates motivation for exercise
- PMID: 36517598
- PMCID: PMC11162758
- DOI: 10.1038/s41586-022-05525-z
A microbiome-dependent gut-brain pathway regulates motivation for exercise
Abstract
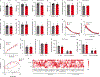
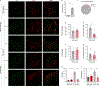
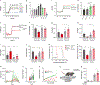
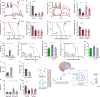
Exercise exerts a wide range of beneficial effects for healthy physiology1. However, the mechanisms regulating an individual's motivation to engage in physical activity remain incompletely understood. An important factor stimulating the engagement in both competitive and recreational exercise is the motivating pleasure derived from prolonged physical activity, which is triggered by exercise-induced neurochemical changes in the brain. Here, we report on the discovery of a gut-brain connection in mice that enhances exercise performance by augmenting dopamine signalling during physical activity. We find that microbiome-dependent production of endocannabinoid metabolites in the gut stimulates the activity of TRPV1-expressing sensory neurons and thereby elevates dopamine levels in the ventral striatum during exercise. Stimulation of this pathway improves running performance, whereas microbiome depletion, peripheral endocannabinoid receptor inhibition, ablation of spinal afferent neurons or dopamine blockade abrogate exercise capacity. These findings indicate that the rewarding properties of exercise are influenced by gut-derived interoceptive circuits and provide a microbiome-dependent explanation for interindividual variability in exercise performance. Our study also suggests that interoceptomimetic molecules that stimulate the transmission of gut-derived signals to the brain may enhance the motivation for exercise.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures











Comment in
-
Gut microbes shape athletic motivation.Nature. 2022 Dec;612(7941):633-634. doi: 10.1038/d41586-022-04355-3. Nature. 2022. PMID: 36517676 No abstract available.
-
Motivation for exercise from the gut.Nat Rev Microbiol. 2023 Mar;21(3):130. doi: 10.1038/s41579-022-00851-5. Nat Rev Microbiol. 2023. PMID: 36600073 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

