Microglia regulate central nervous system myelin growth and integrity
- PMID: 36517604
- PMCID: PMC9812791
- DOI: 10.1038/s41586-022-05534-y
Microglia regulate central nervous system myelin growth and integrity
Erratum in
-
Author Correction: Microglia regulate central nervous system myelin growth and integrity.Nature. 2024 Jul;631(8021):E11. doi: 10.1038/s41586-024-07696-3. Nature. 2024. PMID: 38961305 Free PMC article. No abstract available.
Abstract
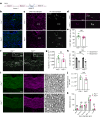
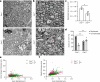
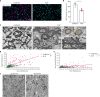
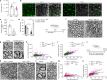
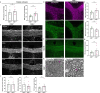
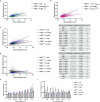
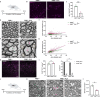
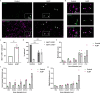
Myelin is required for the function of neuronal axons in the central nervous system, but the mechanisms that support myelin health are unclear. Although macrophages in the central nervous system have been implicated in myelin health1, it is unknown which macrophage populations are involved and which aspects they influence. Here we show that resident microglia are crucial for the maintenance of myelin health in adulthood in both mice and humans. We demonstrate that microglia are dispensable for developmental myelin ensheathment. However, they are required for subsequent regulation of myelin growth and associated cognitive function, and for preservation of myelin integrity by preventing its degeneration. We show that loss of myelin health due to the absence of microglia is associated with the appearance of a myelinating oligodendrocyte state with altered lipid metabolism. Moreover, this mechanism is regulated through disruption of the TGFβ1-TGFβR1 axis. Our findings highlight microglia as promising therapeutic targets for conditions in which myelin growth and integrity are dysregulated, such as in ageing and neurodegenerative disease2,3.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

