Assessment of models for calculating the hydrodynamic radius of intrinsically disordered proteins
- PMID: 36518077
- PMCID: PMC9892621
- DOI: 10.1016/j.bpj.2022.12.013
Assessment of models for calculating the hydrodynamic radius of intrinsically disordered proteins
Abstract
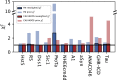
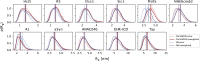
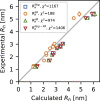
Diffusion measurements by pulsed-field gradient NMR and fluorescence correlation spectroscopy can be used to probe the hydrodynamic radius of proteins, which contains information about the overall dimension of a protein in solution. The comparison of this value with structural models of intrinsically disordered proteins is nonetheless impaired by the uncertainty of the accuracy of the methods for computing the hydrodynamic radius from atomic coordinates. To tackle this issue, we here build conformational ensembles of 11 intrinsically disordered proteins that we ensure are in agreement with measurements of compaction by small-angle x-ray scattering. We then use these ensembles to identify the forward model that more closely fits the radii derived from pulsed-field gradient NMR diffusion experiments. Of the models we examined, we find that the Kirkwood-Riseman equation provides the best description of the hydrodynamic radius probed by pulsed-field gradient NMR experiments. While some minor discrepancies remain, our results enable better use of measurements of the hydrodynamic radius in integrative modeling and for force field benchmarking and parameterization.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. http://www.nature.com/articles/nrm3920 - PMC - PubMed
-
- Orioli S., Larsen A.H., et al. Lindorff-Larsen K. In: Computational Approaches for Understanding Dynamical Systems: Protein Folding and Assembly. Strodel B., Barz B., editors. Academic Press; 2020. Chapter Three - how to learn from inconsistencies: integrating molecular simulations with experimental data; pp. 123–176.https://www.sciencedirect.com/science/article/pii/S1877117319302121 Volume 170 of Progress in Molecular Biology and Translational Science. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

