A Prognostic Model of Seven Immune Genes to Predict Overall Survival in Childhood Acute Myeloid Leukemia
- PMID: 36518627
- PMCID: PMC9744619
- DOI: 10.1155/2022/7724220
A Prognostic Model of Seven Immune Genes to Predict Overall Survival in Childhood Acute Myeloid Leukemia
Abstract
Background: Acute myeloid leukemia (AML) is one of the most common hematological malignancies and accounts for about 20% of childhood leukemias. Currently, immunotherapy is one of the recommended treatment schemes for recurrent AML patients to improve their survival rates. Nonetheless, low remission and high mortality rates are observed in recurrent AML and challenge the prognosis of AML patients. To address this problem, we aimed to establish and verify a reliable prognostic risk model using immune-related genes to improve the prognostic evaluation and recommendation for personalized treatment of AML.
Methods: Transcriptome data and clinical data were acquired from the TARGET database while immune genes were sourced from InnateDB and ImmPort Shared databases. The mRNA expression profile matrix of immune genes from 62 normal samples and 1408 AML cases was extracted from the transcriptome data and subjected to differential expression (DE) analysis. The entire cohort of DE immune genes was randomly divided into the test group and training group. The prognostic model associated with immune genes was constructed using the training group. The test group and entire cohort were employed for model validation. Lastly, we analyzed the potential clinical application of the model and its association with immune cell infiltration.
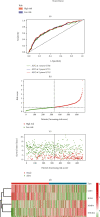
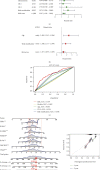
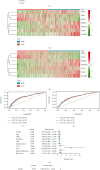
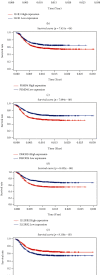
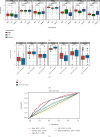
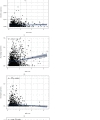
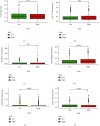
Results: In total, 751 DE immune genes were differentially regulated, including 552 upregulated and 199 downregulated. Based on these DE genes, we developed and validated a prognostic risk model composed of seven immune genes, GDF1, TPM2, IL1R1, PSMD4, IL5RA, DHCR24, and IL12RB2. This model is able to predict the 5-year survival rate more accurately compared with age, gender, and risk stratification. Further analysis showed that CD8+ T-cell contents and neutrophil infiltration decreased but macrophage infiltration increased as the risk score increased.
Conclusions: A seven-immune gene model of AML was developed and validated. We propose this model as an independent prognostic variable able to estimate the 5-year survival rate. In addition, the model can also reflect the immune microenvironment of AML patients.
Copyright © 2022 Yan Luo et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this manuscript.
Figures

References
-
- Moerloose B., Reedijk A., Bock G. H., et al. Response-guided chemotherapy for pediatric acute myeloid leukemia without hematopoietic stem cell transplantation in first complete remission: results from protocol DB AML-01. Pediatric Blood & Cancer . 2019;66(5, article e27605) doi: 10.1002/pbc.27605. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

