Longer epidermal cells underlie a quantitative source of variation in wheat flag leaf size
- PMID: 36519272
- PMCID: PMC10107444
- DOI: 10.1111/nph.18676
Longer epidermal cells underlie a quantitative source of variation in wheat flag leaf size
Abstract
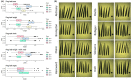
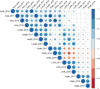
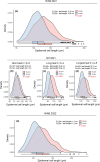
The wheat flag leaf is the main contributor of photosynthetic assimilates to developing grains. Understanding how canopy architecture strategies affect source strength and yield will aid improved crop design. We used an eight-founder population to investigate the genetic architecture of flag leaf area, length, width and angle in European wheat. For the strongest genetic locus identified, we subsequently created a near-isogenic line (NIL) pair for more detailed investigation across seven test environments. Genetic control of traits investigated was highly polygenic, with colocalisation of replicated quantitative trait loci (QTL) for one or more traits identifying 24 loci. For QTL QFll.niab-5A.1 (FLL5A), development of a NIL pair found the FLL5A+ allele commonly conferred a c. 7% increase in flag and second leaf length and a more erect leaf angle, resulting in higher flag and/or second leaf area. Increased FLL5A-mediated flag leaf length was associated with: (1) longer pavement cells and (2) larger stomata at lower density, with a trend for decreased maximum stomatal conductance (Gsmax ) per unit leaf area. For FLL5A, cell size rather than number predominantly determined leaf length. The observed trade-offs between leaf size and stomatal morphology highlight the need for future studies to consider these traits at the whole-leaf level.
Keywords: flag leaf morphology; haplotype analysis; maximum stomatal conductance (Gsmax); multifounder advanced generation intercross population; quantitative trait variation; wheat (Triticum aestivum L.).
© 2022 The Authors New Phytologist © 2022 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures





References
-
- Altschul SF, Gish W, Miller E, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. - PubMed
-
- Austin R, Ford M, Edrich J, Hooper B. 1976. Some effects of leaf posture on photosynthesis and yield in wheat. Annals of Applied Biology 83: 425–446.
-
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 15: 263–265. - PubMed
-
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed‐effects models using lme4. Journal of Statistical Software 67: 1–48.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

