UncertainSCI: Uncertainty quantification for computational models in biomedicine and bioengineering
- PMID: 36521358
- PMCID: PMC9812870
- DOI: 10.1016/j.compbiomed.2022.106407
UncertainSCI: Uncertainty quantification for computational models in biomedicine and bioengineering
Abstract
Background: Computational biomedical simulations frequently contain parameters that model physical features, material coefficients, and physiological effects, whose values are typically assumed known a priori. Understanding the effect of variability in those assumed values is currently a topic of great interest. A general-purpose software tool that quantifies how variation in these parameters affects model outputs is not broadly available in biomedicine. For this reason, we developed the 'UncertainSCI' uncertainty quantification software suite to facilitate analysis of uncertainty due to parametric variability.
Methods: We developed and distributed a new open-source Python-based software tool, UncertainSCI, which employs advanced parameter sampling techniques to build polynomial chaos (PC) emulators that can be used to predict model outputs for general parameter values. Uncertainty of model outputs is studied by modeling parameters as random variables, and model output statistics and sensitivities are then easily computed from the emulator. Our approaches utilize modern, near-optimal techniques for sampling and PC construction based on weighted Fekete points constructed by subsampling from a suitably randomized candidate set.
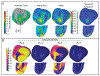
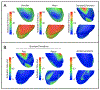
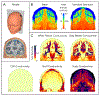
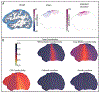
Results: Concentrating on two test cases-modeling bioelectric potentials in the heart and electric stimulation in the brain-we illustrate the use of UncertainSCI to estimate variability, statistics, and sensitivities associated with multiple parameters in these models.
Conclusion: UncertainSCI is a powerful yet lightweight tool enabling sophisticated probing of parametric variability and uncertainty in biomedical simulations. Its non-intrusive pipeline allows users to leverage existing software libraries and suites to accurately ascertain parametric uncertainty in a variety of applications.
Keywords: Biomedical simulations; Open-source software; Uncertainty quantification.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Adams BM, Bohnhoff WJ, Dalbey KR, Ebeida MS, Eddy JP, Eldred MS, Hooper RW, Hough PD, Hu KT, Jakeman JD, Khalil M, Maupin KA, Monschke JA, Ridgeway EM, Rushdi AA, Seidl DT, Stephens JA, Swiler LP, and Winokur JG, DAKOTA, a multilevel parallel object-oriented framework for design optimization, parameter estimation, uncertainty quantification, and sensitivity analysis: Version 6.15 User’s Manual, Tech. report, Sandia National Laboratories Albuquerque, NM, 2021, Sandia Technical Report SAND2020-12495.
-
- Bos L, De Marchi S, Sommariva A, and Vianello M, Computing Multivariate Fekete and Leja Points by Numerical Linear Algebra, SIAM Journal on Numerical Analysis 48 (2010), no. 5, 1984.
-
- Burk Kyle M., Narayan Akil, and Orr Joseph A., Efficient sampling for polynomial chaos-based uncertainty quantification and sensitivity analysis using weighted approximate Fekete points, International Journal for Numerical Methods in Biomedical Engineering 36 (2020), no. 11, e3395, arXiv: 2008.04854. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

