Dysfunctional Extracellular Matrix Remodeling Supports Perianal Fistulizing Crohn's Disease by a Mechanoregulated Activation of the Epithelial-to-Mesenchymal Transition
- PMID: 36521659
- PMCID: PMC9898761
- DOI: 10.1016/j.jcmgh.2022.12.006
Dysfunctional Extracellular Matrix Remodeling Supports Perianal Fistulizing Crohn's Disease by a Mechanoregulated Activation of the Epithelial-to-Mesenchymal Transition
Abstract
Background and aims: Perianal fistula represents one of the most disabling manifestations of Crohn's disease (CD) due to complete destruction of the affected mucosa, which is replaced by granulation tissue and associated with changes in tissue organization. To date, the molecular mechanisms underlying perianal fistula formation are not well defined. Here, we dissected the tissue changes in the fistula area and addressed whether a dysregulation of extracellular matrix (ECM) homeostasis can support fistula formation.
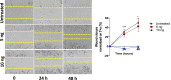
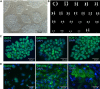
Methods: Surgical specimens from perianal fistula tissue and the surrounding region of fistulizing CD were analyzed histologically and by RNA sequencing. Genes significantly modulated were validated by real-time polymerase chain reaction, Western blot, and immunofluorescence assays. The effect of the protein product of TNF-stimulated gene-6 (TSG-6) on cell morphology, phenotype, and ECM organization was investigated with endogenous lentivirus-induced overexpression of TSG-6 in Caco-2 cells and with exogenous addition of recombinant human TSG-6 protein to primary fibroblasts from region surrounding fistula. Proliferative and migratory assays were performed.
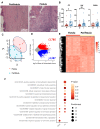
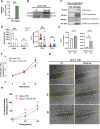
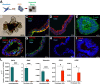
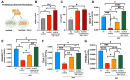
Results: A markedly different organization of ECM was found across fistula and surrounding fistula regions with an increased expression of integrins and matrix metalloproteinases and hyaluronan (HA) staining in the fistula, associated with increased newly synthesized collagen fibers and mechanosensitive proteins. Among dysregulated genes associated with ECM, TNFAI6 (gene encoding for TSG-6) was as significantly upregulated in the fistula compared with area surrounding fistula, where it promoted the pathological formation of complexes between heavy chains from inter-alpha-inhibitor and HA responsible for the formation of a crosslinked ECM. There was a positive correlation between TNFAI6 expression and expression of mechanosensitive genes in fistula tissue. The overexpression of TSG-6 in Caco-2 cells promoted migration, epithelial-mesenchymal transition, transcription factor SNAI1, and HA synthase (HAs) levels, while in fibroblasts, isolated from the area surrounding the fistula, it promoted an activated phenotype. Moreover, the enrichment of an HA scaffold with recombinant human TSG-6 protein promoted collagen release and increase of SNAI1, ITGA4, ITGA42B, and PTK2B genes, the latter being involved in the transduction of responses to mechanical stimuli.
Conclusions: By mediating changes in the ECM organization, TSG-6 triggers the epithelial-mesenchymal transition transcription factor SNAI1 through the activation of mechanosensitive proteins. These data point to regulators of ECM as new potential targets for the treatment of CD perianal fistula.
Keywords: Crohn′s Disease; EMT; Extracellular Matrix; Perianal Fistula; TSG-6.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






Comment in
-
Unravelling the Matrix in Crohn's Disease.Cell Mol Gastroenterol Hepatol. 2023;15(3):801-802. doi: 10.1016/j.jcmgh.2022.12.007. Epub 2022 Dec 28. Cell Mol Gastroenterol Hepatol. 2023. PMID: 36586719 Free PMC article. No abstract available.
References
-
- Torres J., Mehandru S., Colombel J.F., et al. Crohn‘s disease. Lancet. 2017;389:1741–1755. - PubMed
-
- Roda G., Chien Ng S., Kotze P.G., et al. Crohn‘s disease. Nat Rev Dis Primers. 2020;6:22. - PubMed
-
- Gecse K.B., Bemelman W., Kamm M.A., et al. A global consensus on the classification, diagnosis and multidisciplinary treatment of perianal fistulising Crohn‘s disease. Gut. 2014;63:1381–1392. - PubMed
-
- Beaugerie L., Seksik P., Nion-Larmurier I., et al. Predictors of Crohn‘s disease. Gastroenterology. 2006;130:650–656. - PubMed
-
- Bell S.J., Williams A.B., Wiesel P., et al. The clinical course of fistulating Crohn‘s disease. Aliment Pharmacol Ther. 2003;17:1145–1151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

