A proof-of-concept study on the genomic evolution of Sars-Cov-2 in molnupiravir-treated, paxlovid-treated and drug-naïve patients
- PMID: 36522489
- PMCID: PMC9753865
- DOI: 10.1038/s42003-022-04322-8
A proof-of-concept study on the genomic evolution of Sars-Cov-2 in molnupiravir-treated, paxlovid-treated and drug-naïve patients
Abstract
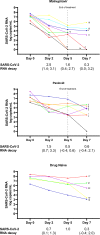
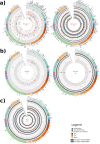
Little is known about SARS-CoV-2 evolution under Molnupiravir and Paxlovid, the only antivirals approved for COVID-19 treatment. By investigating SARS-CoV-2 variability in 8 Molnupiravir-treated, 7 Paxlovid-treated and 5 drug-naïve individuals at 4 time-points (Days 0-2-5-7), a higher genetic distance is found under Molnupiravir pressure compared to Paxlovid and no-drug pressure (nucleotide-substitutions/site mean±Standard error: 18.7 × 10-4 ± 2.1 × 10-4 vs. 3.3 × 10-4 ± 0.8 × 10-4 vs. 3.1 × 10-4 ± 0.8 × 10-4, P = 0.0003), peaking between Day 2 and 5. Molnupiravir drives the emergence of more G-A and C-T transitions than other mutations (P = 0.031). SARS-CoV-2 selective evolution under Molnupiravir pressure does not differ from that under Paxlovid or no-drug pressure, except for orf8 (dN > dS, P = 0.001); few amino acid mutations are enriched at specific sites. No RNA-dependent RNA polymerase (RdRp) or main proteases (Mpro) mutations conferring resistance to Molnupiravir or Paxlovid are found. This proof-of-concept study defines the SARS-CoV-2 within-host evolution during antiviral treatment, confirming higher in vivo variability induced by Molnupiravir compared to Paxlovid and drug-naive, albeit not resulting in apparent mutation selection.
© 2022. The Author(s).
Conflict of interest statement
We have no competing interest regarding the data reported in this paper. As general competing interests, C.F.P. acknowledges grants, boards, and sponsored lectures from Gilead, ViiV, Merck, Janssen, GSK, Astra Zeneca. C.M. has received research grants from Gilead and participated to advisory boards for Gilead, ViiV, Janssen, MSD, Angelini, Roche. C.A. acknowledges sponsored lectures from Pfizer. A.V. acknowledges research grants from MSD. S.B. has received research grants from Gilead. C.R. acknowledges research grants from DiaSorin. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

