Mitochondrial genomes assembled from non-invasive eDNA metagenomic scat samples in the endangered Amur tiger Panthera tigris altaica
- PMID: 36523460
- PMCID: PMC9745948
- DOI: 10.7717/peerj.14428
Mitochondrial genomes assembled from non-invasive eDNA metagenomic scat samples in the endangered Amur tiger Panthera tigris altaica
Abstract
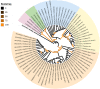
The Amur or Siberian tiger Panthera tigris altaica (Temminck, 1844) is currently restricted to a small region of its original geographical range in northwestern Asia and is considered 'endangered' by the IUCN Red List of Threatened Species. This solitary, territorial, and large top predator is in major need of genomic resources to inform conservation management strategies. This study formally tested if complete mitochondrial genomes of P. tigris altaica can be assembled from non-enriched metagenomic libraries generated from scat eDNA samples using the Illumina sequencing platform and open-access bioinformatics pipelines. The mitogenome of P. tigris altaica was assembled and circularized using the pipeline GetOrganelle with a coverage ranging from 322.7x to 17.6x in four different scat eDNA samples. A nearly complete mitochondrial genome (101x) was retrieved from a fifth scat eDNA sample. The complete or nearly complete mitochondrial genomes of P. tigris altaica were AT-rich and composed of 13 protein coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes, and a putative control region. Synteny observed in all assembled mitogenomes was identical to that reported before for P. tigris altaica and other felids. A phylogenomic analysis based on all PCGs demonstrated that the mitochondrial genomes assembled from scat eDNA reliably identify the sequenced samples as belonging to P. tigris and distinguished the same samples from closely and distantly related congeneric species. This study demonstrates that it is viable to retrieve accurate whole and nearly complete mitochondrial genomes of P. tigris altaica (and probably other felids) from scat eDNA samples without library enrichment protocols and using open-access bioinformatics workflows. This new genomic resource represents a new tool to support conservation strategies (bio-prospecting and bio-monitoring) in this iconic cat.
Keywords: Endangered species; Metagenome; Mitochondrial genomes; eDNA.
© 2022 Baeza.
Conflict of interest statement
The author declares that they have no competing interests.
Figures


References
-
- Barnett R, Mendoza MLZ, Soares AER, Ho SY, Zazula G, Yamaguchi N, Shapiro B, Kirillova IV, Larson G, Gilbert MTP. Mitogenomics of the extinct cave lion, Panthera spelaea (Goldfuss, 1810), resolve its position within the Panthera cats. Open Quaternary. 2016;2:1–11. doi: 10.5334/oq.24. - DOI
-
- Bozarth CA, Alva-Campbell YR, Ralls K, Henry TR, Smith DA, Westphal MF, Maldonado JE. An efficient noninvasive method for discriminating among faeces of sympatric North American canids. Conservation Genetics Resources. 2010;2(1):173–175. doi: 10.1007/s12686-010-9215-4. - DOI
-
- Bradshaw CJ, Ehrlich PR, Beattie A, Ceballos G, Crist E, Diamond J, Dirzo R, Ehrlich AH, Harte J, Harte ME, Pyke G. Underestimating the challenges of avoiding a ghastly future. Frontiers in Conservation Science. 2021;1:615419. doi: 10.3389/fcosc.2020.615419. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

