Combined proteomics and single cell RNA-sequencing analysis to identify biomarkers of disease diagnosis and disease exacerbation for systemic lupus erythematosus
- PMID: 36524113
- PMCID: PMC9746895
- DOI: 10.3389/fimmu.2022.969509
Combined proteomics and single cell RNA-sequencing analysis to identify biomarkers of disease diagnosis and disease exacerbation for systemic lupus erythematosus
Abstract
Introduction: Systemic lupus erythematosus (SLE) is a chronic autoimmune disease for which there is no cure. Effective diagnosis and precise assessment of disease exacerbation remains a major challenge.
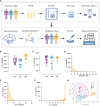
Methods: We performed peripheral blood mononuclear cell (PBMC) proteomics of a discovery cohort, including patients with active SLE and inactive SLE, patients with rheumatoid arthritis (RA), and healthy controls (HC). Then, we performed a machine learning pipeline to identify biomarker combinations. The biomarker combinations were further validated using enzyme-linked immunosorbent assays (ELISAs) in another cohort. Single-cell RNA sequencing (scRNA-seq) data from active SLE, inactive SLE, and HC PBMC samples further elucidated the potential immune cellular sources of each of these PBMC biomarkers.
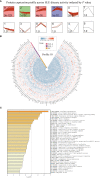
Results: Screening of the PBMC proteome identified 1023, 168, and 124 proteins that were significantly different between SLE vs. HC, SLE vs. RA, and active SLE vs. inactive SLE, respectively. The machine learning pipeline identified two biomarker combinations that accurately distinguished patients with SLE from controls and discriminated between active and inactive SLE. The validated results of ELISAs for two biomarker combinations were in line with the discovery cohort results. Among them, the six-protein combination (IFIT3, MX1, TOMM40, STAT1, STAT2, and OAS3) exhibited good performance for SLE disease diagnosis, with AUC of 0.723 and 0.815 for distinguishing SLE from HC and RA, respectively. Nine-protein combination (PHACTR2, GOT2, L-selectin, CMC4, MAP2K1, CMPK2, ECPAS, SRA1, and STAT2) showed a robust performance in assessing disease exacerbation (AUC=0.990). Further, the potential immune cellular sources of nine PBMC biomarkers, which had the consistent changes with the proteomics data, were elucidated by PBMC scRNAseq.
Discussion: Unbiased proteomic quantification and experimental validation of PBMC samples from two cohorts of patients with SLE were identified as biomarker combinations for diagnosis and activity monitoring. Furthermore, the immune cell subtype origin of the biomarkers in the transcript expression level was determined using PBMC scRNAseq. These findings present valuable PBMC biomarkers associated with SLE and may reveal potential therapeutic targets.
Keywords: biomarker; disease diagnosis; disease exacerbation; immune cell; machine learning.
Copyright © 2022 Li, Ma, Liao, Qi, Meng, Cai, Dai, Cao, Dong, Krämer, Yun, Hocher, Hong, Liu, Tang, He, Yin and Dai.
Conflict of interest statement
Author LY was employed by Guangzhou Enttxs Medical Products Co., Ltd. Author BH was employed by Reproductive and Genetic Hospital of China International Trust and Investment Corporation (CITIC)-Xiangya. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Enocsson H, Wirestam L, Dahle C, Padyukov L, Jönsen A, Urowitz MB, et al. . Soluble urokinase plasminogen activator receptor (Supar) levels predict damage accrual in patients with recent-onset systemic lupus erythematosus. J Autoimmun (2020) 106:102340. doi: 10.1016/j.jaut.2019.102340 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

